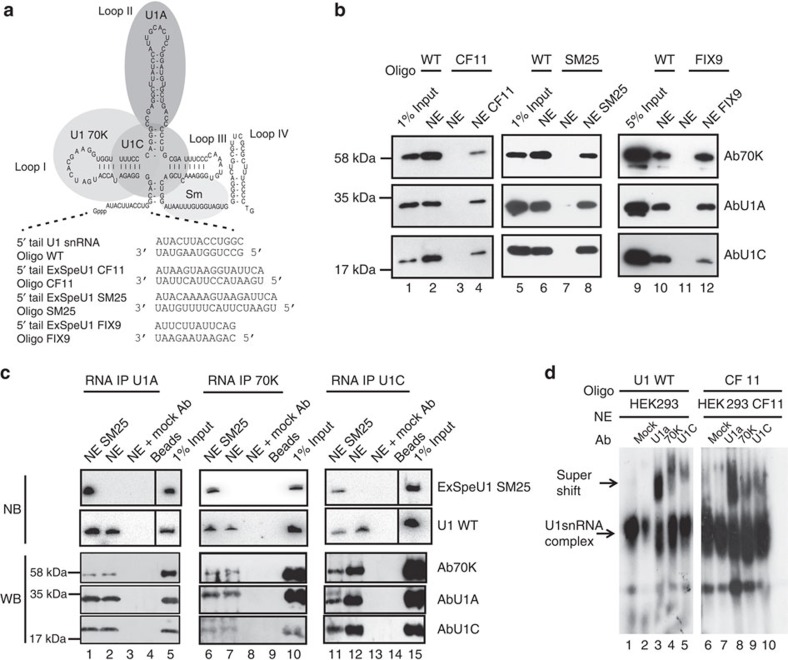
Figure 4. Protein composition of ExSpeU1s and endogenous U1 snRNP.
(a) Schematic representation of U1 snRNA secondary structure and associated proteins along with the RNA oligonucleotides used in affinity purification, RIP and EMSA. (b) Affinity purified CF11, SM25 and FIX9 ExSpeU1s contain 70K, U1A and U1C. Nuclear extracts (NE) from Hek293 cells, transfected with the indicated ExSpeU1s, or not transfected cells were incubated with the corresponding biotinylated 2′-O-methyl-RNA oligonucleotides. Affinity-purified snRNPs were analysed by western blotting using antibodies against U1–70K, U1A and U1C. (c) RNA-immunoprecipitation analysis of SM25 ExSpeU1. Hek293 NEs from cells transfected with ExSpeU1 SM25 or not transfected cells were incubated with antibodies against U1A, 70K and U1C. RNAs and proteins purified from the RIP complexes were analysed by northern and western blotting, respectively with the indicated probes/antibodies. Mock Ab corresponds to anti-tubulin. (d) CF11 ExSpeU1 snRNP has the same electrophoretic mobility as normal U1 in EMSA. Radiolabelled RNA oligonucleotides complementary to normal U1 (lanes 1–5) or ExSpeU1 CF11 (lanes 6–10) were incubated with nuclear extracts transfected with ExSpe CF11 or mock (not transfected cells). Addition of the indicated antibodies super-shifted the complexes in U1wt and ExSpe CF11. Control EMSA experiments are shown in Supplementary Fig. 5.