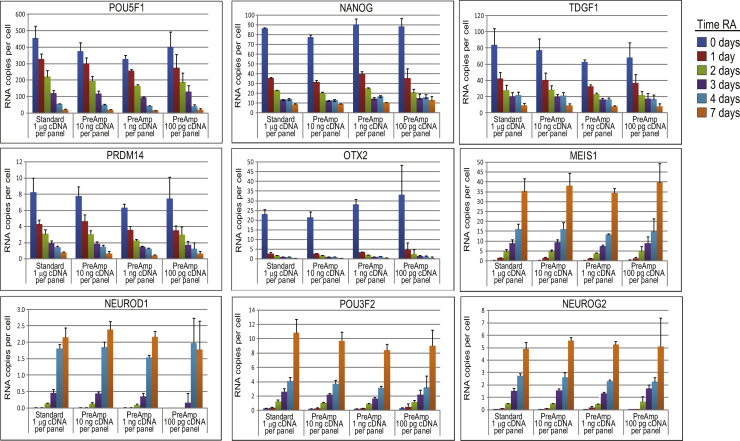
Fig. 2.
Fold-change bias of native target genes with limited sample. cDNA derived from NT2 cells treated with RA for 0–7 days were pre-amplified for 10 PCR cycles and used to quantify native target genes by qPCR; 100 pg, 1 ng and 10 ng cDNA was used in the PreAmp reactions. 1 μg cDNA that was not pre-amplified was also used to quantify target gene expression by standard qPCR (10 ng per target). The level of expression of each target gene was determined and expressed as “RNA copies per cell” after normalizing with TBP and assuming that there are 10 TBP copies per cell. The experiment was performed three times, each with two technical replicates, the average values are reported. Error bars represent standard deviation. Data for selected target genes that show RA-induced changes are shown.