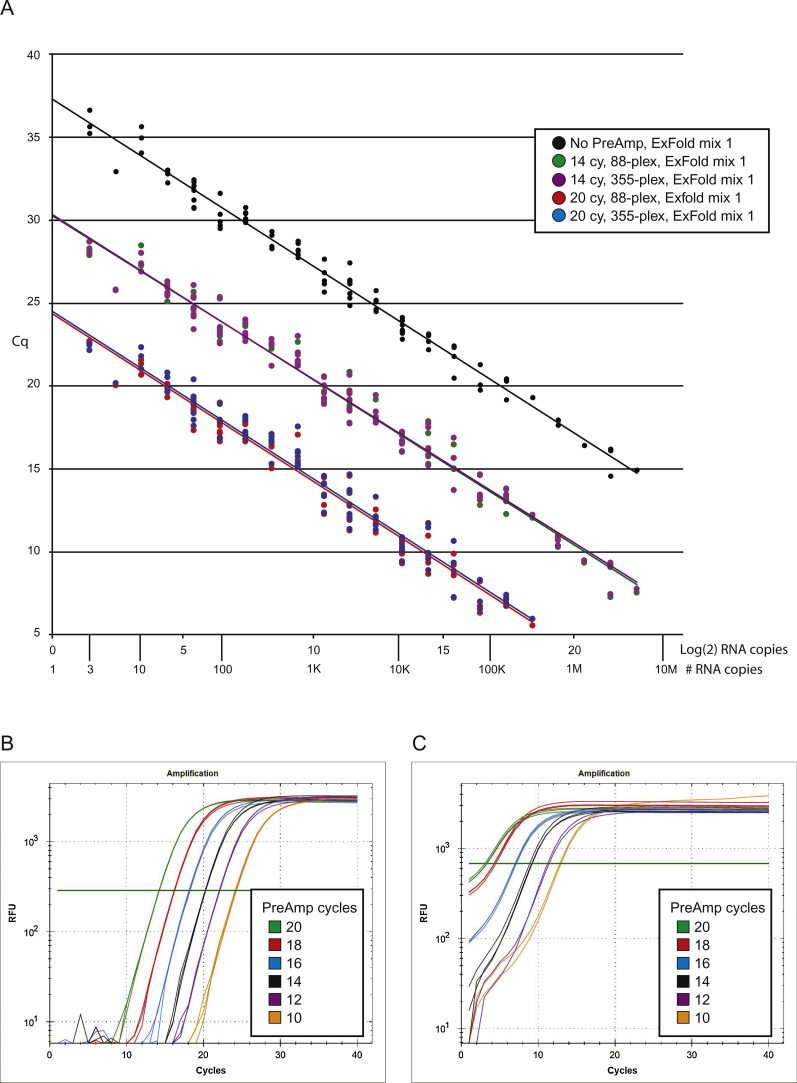
Fig. 4.
Dynamic range bias. ExFold spike in-mixtures were added to 10 ng NT2 RNA, reverse transcribed and pre-amplified for 10–20 PCR cycles; ERCC targets were quantified by qPCR using two technical replicates, the experiment was performed one time. (A) The measured amount of ERCC target gene is plotted as a function of actual RNA abundance. All ERCC targets can be detected in the no PreAmp sample and in the samples processed for 14 PreAmp cycles. For samples processed for 20 PreAmp cycles, quantification of targets in the 1 million copy range, have a Cq value less than 5 and are of unreliable value. (B) qPCR results for ERCC-00144, which contained 5,513 copies in the PreAmp reaction, show good results over the entire amplification range. (C) qPCR results for ERCC-00074, which contained 2,822,879 copies in the PreAmp reaction, show poor results when amplified for 18 or 20 PCR cycles.