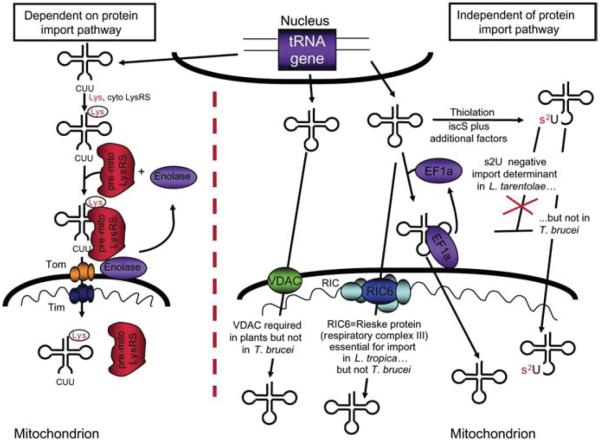
Figure 1.
Two major pathways for tRNA import into mitochondria.
One pathway utilizes the protein import machinery including the translocases of the outer membrane (TOMs) and inner membrane (TIMs). This pathway has thus far only been described in the import of tRNALysCUU in yeast and is depicted in the left side of the Figure. In this pathway, the imported tRNA is first aminoacylated by its cognate cytoplasmic synthetase (cyto-LysRS), bound by the pre-mitochondrial lysyl tRNA synthetase (pre-mitoLysRS) and with the aid of enolase is delivered to the mitochondrial surface. The complex is then translocated via the protein import machinery, while enolase remains in the cytoplasm where it can start a new import cycle. The second and perhaps phylogenetically the most widely distributed pathway does not require cytoplasmic factors and it is not directly dependent on the protein import machinery. However, various factors may play a role in various organisms including the VDAC (plants), tRNA thiolation (Leishmania but not in T. brucei), etc. In the specific case of T. brucei, the translation elongation factor eEF1a plays a role in delivering the tRNA to the mitochondrial import machinery. This role is similar to that of enolase in yeast, but has yet to be demonstrated in other organisms. In L. tropica, the RNA import complex (RIC) was recently described and is formed by six essential subunits, all are identical to subunits of the respiratory complex. In T. brucei, however, at least one of the essential RIC subunits is not involved in tRNA import; therefore, the pathway may differ between the two organisms despite being evolutionarily closely related.