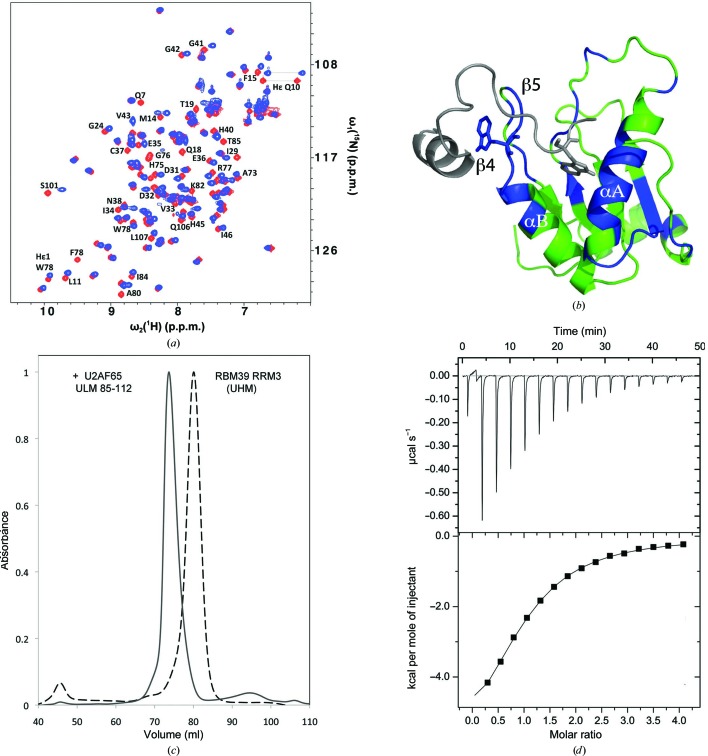
Figure 3.
Biochemical analysis of RBM39-UHM–U2AF65-ULM interactions. (a) Superposition of the two-dimensional [15N,1H]-HSQC spectra of 15N-labeled RBM39-UHM in the absence (red) and presence (blue) of 1.3 equivalents of unlabeled U2AF65-ULM (residues 85–112). The resonances of residues with chemical shift changes of ≥0.13 p.p.m. are labeled. (b) Model of the RBM39-UHM–U2AF65-ULM complex generated by HADDOCK. The NMR structure of RBM39-UHM (color-coded as described below) was used as the input, and the U2AF65-ULM fragment (gray) was docked to it using NMR constraints derived from the chemical shift mapping. RBM39-UHM residues experiencing either large chemical shifts, line broadening and chemical shifts, or broadening beyond detection are colored blue, while those with no significant changes are colored green. The reciprocal tryptophans are shown as stick diagrams. (c) Normalized size-exclusion chromatography elution profiles of RBM39-UHM and a mixture of RBM39-UHM and U2AF65-ULM (residues 85–112) in a 1:2 molar ratio. (d) Isothermal titration calorimetry profile of solutions of RBM39-UHM and the U2AF65-ULM (85–112) peptide, showing binding with a K d of 20 µM.