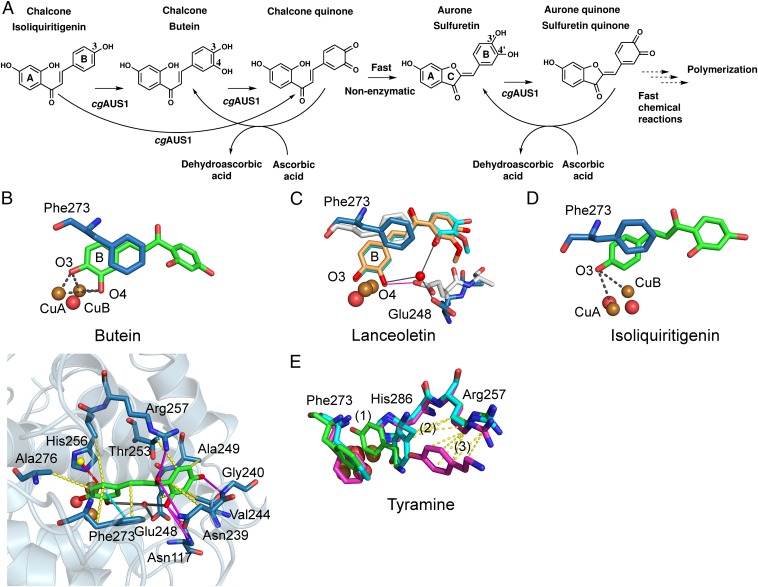
Fig. 4.
Reactivity and in silico-obtained substrate–enzyme complexes of AUS1. (A) Reaction scheme of the hydroxylation of isoliquiritigenin in the presence of ascorbic acid to trap highly reactive quinoid intermediates and products. (B) MD simulation snapshot of AUS1 in complex with butein. The reactive oxygen atoms at the B-ring of butein (O3, O4) are almost symmetrically located between CuA and CuB. The enzyme–substrate interactions are visualized by magenta lines (hydrogen bonding), yellow dashes (hydrophobic interactions), cyan dashed lines (π–π stacking), red dashes (cation–π interactions), and gray lines (water bridges). (C) Superimposition of snapshots obtained from different MD simulations of AUS1 with lanceoletin as the substrate. Residue Glu248 coordinates an oxygen atom of the A-ring through the formation of a water bridge (blue sticks; water bridges, gray lines) or one reactive oxygen atom (O4, B-ring) directly (white sticks; hydrogen bonding, magenta lines). (D) MD simulation snapshot of AUS1 in complex with isoliquiritigenin. (E) Three snapshots [0 ps, green (1); 44 ps, cyan (2); and 344 ps, magenta (3)] of the MD simulation of AUS1 with tyramine. Hydrophobic interactions of tyramine with Arg257 are visualized by yellow dashes.