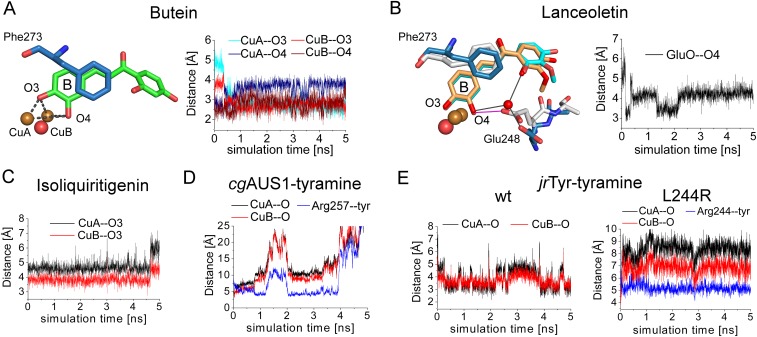
Fig. S6.
Distance plots of the performed MD simulations. (A) Distances of the reactive oxygen atoms of butein (O3 and O4) to both copper atoms of AUS1. (B) Distance of the carboxylic oxygen atom to the substrate’s copper-coordinating oxygen atom. (C) Distances of the reactive oxygen atom of isoliquiritigenin to the CuA and CuB atoms of AUS1. (D) Distances of the reactive oxygen atom of tyramine to the CuA and CuB atoms of AUS1. The centers of masses were used to determine the distance of tyramine to the side chain of Arg257 (blue line). (E) Distances of the reactive oxygen atom of tyramine to the CuA and CuB atoms of the wild type (Left) and the L244R mutant (Right) of jrTYR. The centers of masses were used to determine the distance of tyramine to the side chain of Arg244 (blue line).