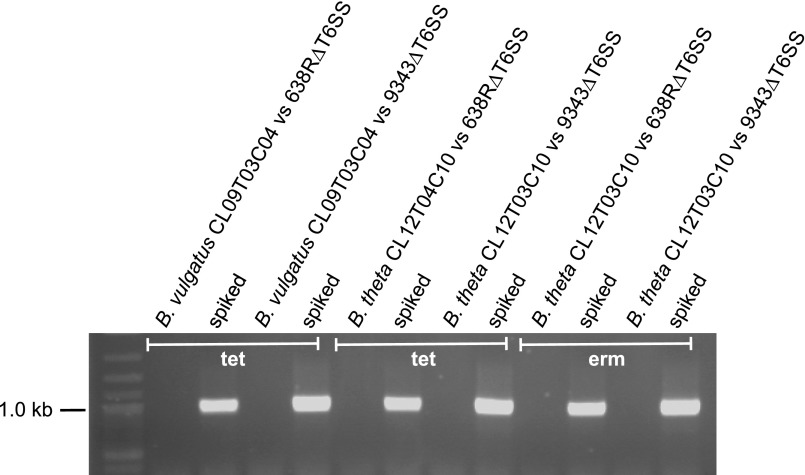
Fig. S1.
Analysis for the presence of predator strain (638R or 9343) on the antibiotic plate following coculture with antibiotic resistant prey strains B. vulgatus CL09T03C04 and B. thetaiotaomicron CL12T03C10. To ensure that the bacteria arising on the erythromycin and/or tetracycline plates following coculture assays were not from the transfer of the antibiotic-resistant gene to the predator strain, the bacteria growing on the third spot from the bottom of cocultures with the T6SS mutants were removed and grown overnight in the antibiotic to ensure dead bacteria were not analyzed. A total of 1 mL of this culture was pelleted, or 1 mL with the addition of 1 µL of 638R or 9343 (spiked). The bacteria were resuspended in 1 mL water, and 1 µL was analyzed by PCR with primers to GA3 conserved regions. No predator bacteria were detected in these samples. The spiked samples show that this method would identify predator bacteria that are present below 1/1,000 of the population if present. B. thetaiotaomicron CL12T03C10 was analyzed for transfer of both the tetracycline and erythromycin resistance genes during coculture.