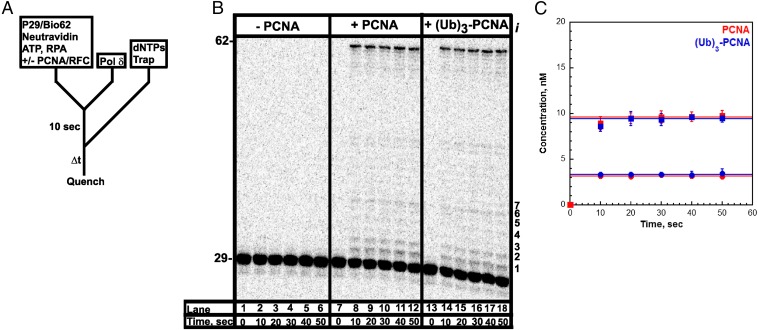
Fig. 2.
Assembly of a pol δ holoenzyme on the P29/Bio62 DNA substrate is characterized by primer extension. (A) Schematic representation of the experiment performed to monitor primer extension by a pol δ holoenzyme. The pol δ holoenzyme was assembled on the P29/Bio62 by the addition of RPA, PCNA, ATP, RFC, and pol δ in succession. PCNA was either unmodified (PCNA) or monoubiquitinated [(Ub)3-PCNA]. Synthesis by assembled holoenzymes was initiated by the simultaneous addition of dNTPs and a passive DNA trap. The experiment for pol δ alone was performed identically except for the omission of PCNA and RFC. (B) 16% denaturing sequencing gel of the primer extension products for pol δ alone (lanes 1–6) and pol δ holoenzymes assembled with either PCNA (lanes 7–12) or (Ub)3-PCNA (lanes 13–18). The sizes of the substrate and full-length product are indicated on the left and the insertion step (i) for each primer extension product up to 36 nt in length (i = 7) is indicated on the right. (C) Quantification of the primer extension products (total, ■) and the fully extended primer (●) for holoenzymes assembled with either PCNA or (Ub)3-PCNA. Each point represents the average ± SD of three independent experiments. For the full-length 62-mer product, all data points were fit to a flat line where the y-intercept reflects the amplitude. The total amount of primer extension products increased up to at most 20 s and plateaued thereafter. Such behavior was independent of the preincubation time (Fig. S3) and was not attributable to limiting concentrations of ATP and/or dNTPs (Fig. S4 A and B) or mis-incorporation of the first nucleotide (Fig. S5). For primer extension products, data points after t = 10 s were fit to a flat line where the y-intercept reflects the amplitude. Values are reported in Table 1.