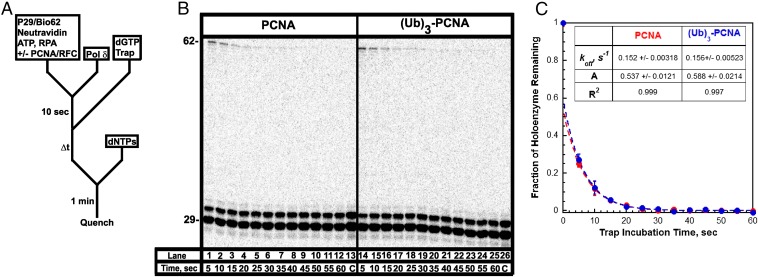
Fig. 4.
Dissociation of a stalled pol δ holoenzyme. (A) Schematic representation of experiment performed to monitor stability of the pol δ holoenzyme. First, the pol δ holoenzyme was assembled as in Fig. 2A. After a 10-s preincubation, trap and dGTP (the first dNTP to be incorporated) were added simultaneously to select for assembled pol δ holoenzymes that are competent for DNA synthesis. After varying incubation times, aliquots were removed, mixed with a dNTP chase containing all dNTPs, and then quenched after 1 min. Under these conditions, only pol δ holoenzymes that remain assembled on the P29/Bio62 substrate after the specified incubation time will be able to extend the primer to i = 2 and beyond. Hence, the probability of insertion for i = 2 is equal to the fraction of pol δ holoenzymes remaining. (B) 16% denaturing sequencing gel of the primer extension products for pol δ holoenzymes assembled with either PCNA (lanes 1–13) or (Ub)3-PCNA (lanes 14–26). The size of the substrate and full-length product is indicated on the left. As a control for each condition, an aliquot was removed after 2 min of preincubation and quenched before the addition of dNTPs (lanes 13 and 26). (C) The fraction of holoenzymes associated with the P29/Bio62 DNA substrate as a function of trap incubation time. Each point represents the average ± SD of three independent experiments for holoenzymes assembled with either PCNA or (Ub)3-PCNA. Data points from 5 to 60 s were fit to a single exponential decay. Extrapolation of the fit back to t = 0 yields the amplitude. The amplitudes, rate constants, and R2 values for each fit are reported.