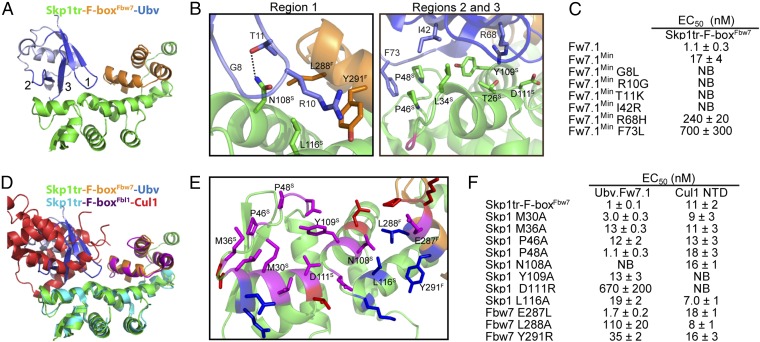
Fig. 2.
Structural and mutational analysis of the interactions between Ubv.Fw7.1 and the Skp1tr–F-boxFbw7 complex. (A) Structure of Ubv.Fw7.1 in complex with Skp1tr–F-boxFbw7. Ubv regions (regions 1–3) that were diversified in Library 1 are labeled and colored dark blue, and other regions are colored light blue. Skp1tr and F-boxFbw7 are colored green or orange, respectively. (B) Details of the molecular interactions between Ubv.Fw7.1 and Skp1tr–F-boxFbw7 showing residues that are mutated relative to WT Ub and are critical for binding. Skp1tr and Fbw7 residues are denoted by “S” and “F” superscripts, respectively. Complex subunits are colored as in A and the location of Loop 1 deleted in Skp1tr is indicated in magenta. (C) Affinities of Ubv.Fw7.1 back-mutants for Skp1tr–F-boxFbw7. Ubv.Fw7.1Min lacks Ub tail (residues 75–78) and contains only six mutations relative to WT Ub (L8G, G10R, K11T, R42I, H68R, and L73F). “NB” indicates no detectable binding. (D) Superposition of Skp1tr–F-boxFbw7-Ubv.Fw7.1 complex with Skp1tr–F-boxFbl1-Cul1 complex (PDB ID code1LDK). Skp1tr–F-boxFbw7–Ubv.Fw7.1 complex subunits are colored as in A and Skp1tr–F-boxFbl1-Cul1 complex subunits are colored as follows: Skp1tr, cyan; F-boxFbl1, purple; Cul1, red. (E) Comparison of the Ubv.Fw7.1-binding and predicted Cul1-binding surfaces on Skp1tr–F-boxFbw7. Skp1tr–F-boxFbw7 residues interacting with Ubv.Fw7.1 or predicted to interact with Cul1 (by comparison with the Skp1–F-boxFbl1–Cul1 complex) are shown as sticks and colored according to predicted interactions: magenta, interacts with Cul1 and Ubv.Fw7.1; red, interacts with Cul1 only; blue, interacts with Ubv.Fw7.1 only. Residues that were subjected to mutagenesis are labeled. (F) Effects of substitutions in Skp1tr or the F-boxFbw7 domain on the binding of Skp1tr–F-boxFbw7 to Ubv.Fw7.1 or Cul1 N-terminal domain (NTD).