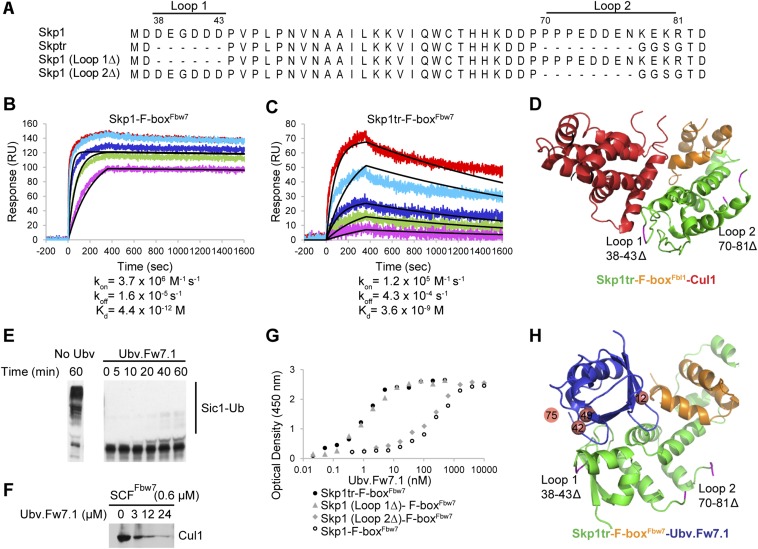
Fig. S2.
Additional in vitro experiments. (A) Alignment of all Skp1 constructs tested in this study. Loop 1 (residues 38–43) and Loop 2 (residues 70–81) indicate residues deleted in truncated version of Skp1 (Skp1tr). (B and C) Binding of Cul1 NTD to Skp1–F-boxFbw7 and Skp1tr–F-boxFbw7 complexes demonstrates importance of Skp1 Loop 1 and Loop 2 residues for interaction with Cul1. Binding was measured by SPR analysis and obtained traces are shown for Cul1 NTD interaction with Skp1-\–F-boxFbw7 (B) and Skp1tr–F-boxFbw7 (C). Skp1–F-box complexes were immobilized and Cul1 NTD was injected at different concentrations. Colored lines represent the actual data (red, 100 nM Cul1; cyan, 33 nM Cul1; blue, 11 nM Cul1; green, 3.7 nM Cul1; and magenta, 1.2 nM Cul1), whereas black lines represent fits to a simple 1:1 Langmuir binding isotherm model. In the case of binding to Skp1–F-boxFbw7, only the three lowest Cul1 concentrations (11 nM Cul1, 3.7 nM Cul1, and 1.2 nM Cul1) were used to fit the data, because higher concentrations exceeded estimated affinity of the interaction >1,000 fold. Estimated kon, koff rates and Kd of the interactions are indicated. (D) Structure of Skp1tr–F-boxFbl1–Cul1 (PDB ID code 1LDK) complex highlighting positions of loops deleted in Skp1tr in relation to Cul1 binding surface. Skp1tr, F-boxFbl1, and Cul1are colored green, orange, or red, respectively. Location of loops deleted in Skp1tr are colored magenta and indicated. (E) Ubv.Fw7.1 blocks ubiquitination activity of SCFFbw7. A functional SCFFbw7complex was assembled from the Skp1tr–(F-box–WD40)Fbw7 and Cul1–Rbx1 complexes purified separately. Sic1, which is a natural substrate of yeast Fbw7 but is also recognized by human Fbw7, was used as the substrate. The ubiquitination reaction containing E1 (0.5 µM), E2 (14 µM), and SCFFbw7 (0.4 µM) was initiated by adding Ub (25 µM) to purified components in the absence and presence of Ubv.Fw7.1 (25 µM). The products of Sic1 (0.4 µM) ubiquitination were visualized by Western blotting at the indicated time points. (F) Ubv.Fw7.1 promotes dissociation of Cul1 from the SCFFbw7 complex. The SCFFbw7 complex containing GST-tagged Skp1tr on glutathionine Sepharose resin was incubated for 1 h with increasing amounts of Ubv.Fw7.1. Cul1 remaining bound to the resin was detected by Western blot. (G) Skp1 Loop 1 (residues 38–43) interferes with binding to Ubv.Fw7.1, whereas Skp1 Loop 2 (residues 70–81) has no effect on binding. Binding was tested by protein ELISA and dose–response curves of Ubv.Fw7.1 binding to different Skp1 constructs in complex with F-boxFbw7 domain are shown. (H) Structure of Skp1tr–F-boxFbw7–Ubv.Fw7.1 complex highlighting positions of loops deleted in Skp1tr. Skp1tr, F-boxFbw7, and Ubv.Fw7.1 are colored green, orange, or blue, respectively. Location of loops deleted in Skp1tr are colored magenta and indicated. Positions that showed different preferences in Ubv selected against Skp1–F-boxFbw7 versus parental Ubv.Fw7.1 sequence are shown as salmon spheres. Position 75 was not defined in the structure and its projected location is shown.