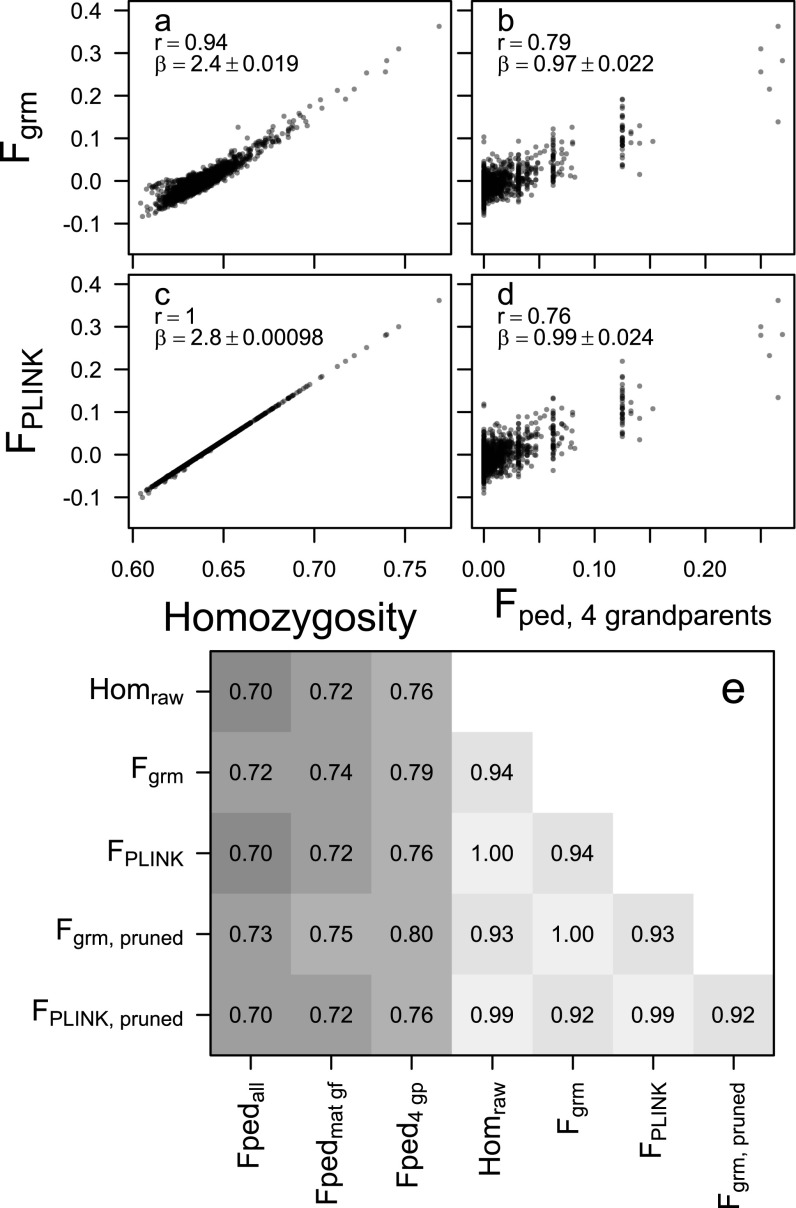
Fig. S2.
Relationships between inbreeding estimated as (A and B) or (C and D) and average SNP homozygosity (A and C, n = 2,248) or pedigree inbreeding for individuals with all four grandparents known (B and D, n = 1,198). Correlations (r) between a wider range of estimators are given in E, with a pairwise sample size of n = 2,248 between all genomic estimators, n = 1944 between genomic estimators and Fpedmat gf (both parents plus maternal grandfather known), and n = 1,198 between genomic estimators and Fped4 gp. “Pruned” refers to using a subset of about 90% of SNPs after pruning on LD (details in SI Materials and Methods).