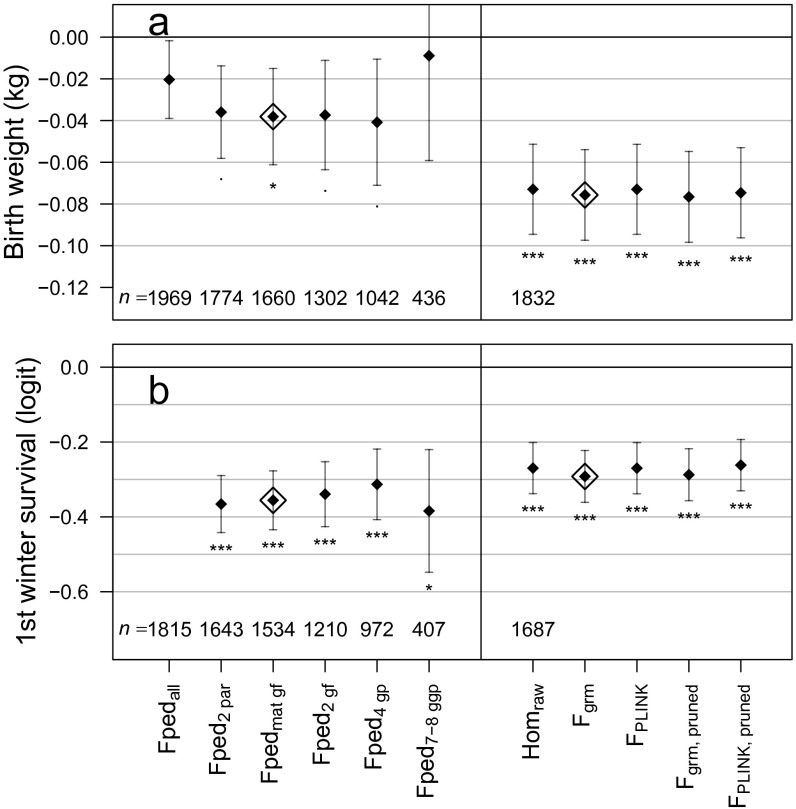
Fig. S4.
Comparison between estimated effect sizes of inbreeding depression using different ancestry thresholds (Left) or different genomic inbreeding estimators (Right) in models for two traits known to show inbreeding depression, birth weight (A) and first winter survival (B; birth weight accounted for). Each estimator was scaled to have an SD of 1 before fitting the model, for this comparison only, and no maternal inbreeding coefficients were fitted; models are otherwise identical to those used in the main text. The pedigree and genomic estimators used in the main text are indicated by open diamonds. Sample sizes are given for each model, and all models with genomic estimators have the same sample size. Asterisks denote standard levels of significance. The subset of individuals with both parents and at least one grandparent known is omitted, as it was identical to the subset with both parents known. The model for first winter survival using including all individuals (Fpedall) failed to converge. “Pruned” refers to using a subset of about 90% of SNPs after pruning on LD (details in SI Materials and Methods).