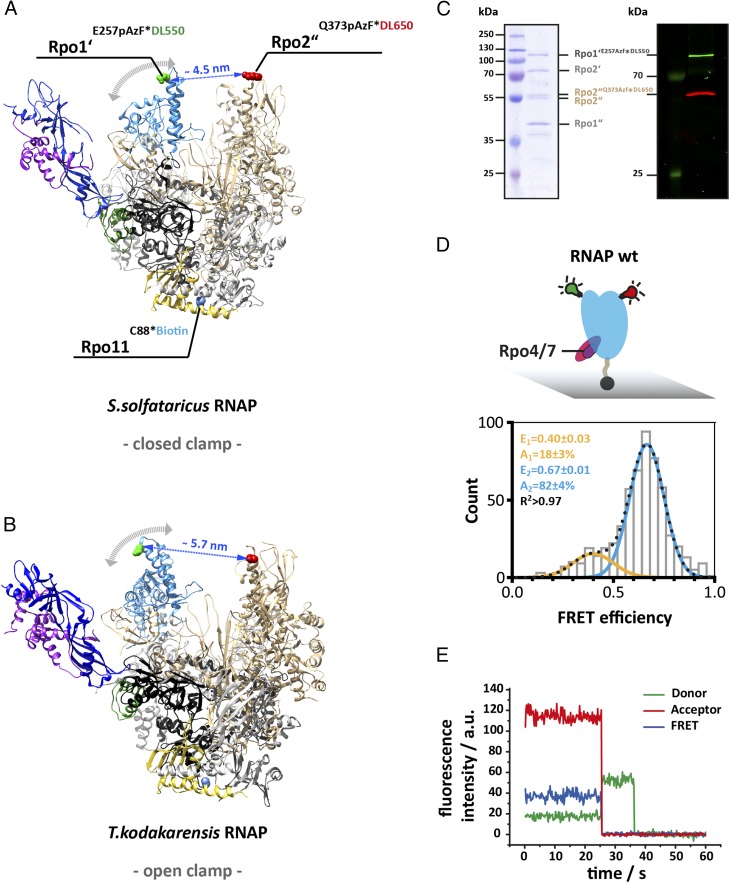
Fig. 1.
Accessing the conformation of the RNA polymerase clamp in solution using single-molecule FRET. Structure of the 12-subunit archaeal RNA polymerase with a (A) closed and (B) open clamp (Protein Data Bank ID codes 2PMZ and 4QIW, respectively). For FRET measurement, a donor (DyLight550) and acceptor (DyLight650) fluorophore were incorporated at the tip of the RNA polymerase clamp coiled coil (subunit Rpo1’ E257, highlighted in green) and the protrusion domain (subunit Rpo2” Q373, highlighted in red). For site-specific labeling the unnatural amino acid p-azidophenylalanine was incorporated at these sites followed by subsequent labeling via Staudinger-Bertozzi ligation. For immobilization purposes a biotin was coupled to a native cysteine in subunit Rpo11. Subunit coloring is as follows: Rpo1′ in dark gray, Rpo2″ in beige, Rpo11 in yellow, Rpo4 in purple, and Rpo7 in blue. (C) The Coomassie 10% SDS/PAGE shows the four large subunits (Left) and the fluorescence scan (Right) of a fully assembled RNAP after purification and demonstrates the specific labeling of subunit Rpo1′ and Rpo2″ with the donor or acceptor dye, respectively. (D) Conformation of the 12-subunit RNA polymerase clamp in the absence of nucleic acid substrates. Biotinylated RNAPs (symbolized by black sphere protruding from the RNAP core) were immobilized on a quartz slide (gray) for TIRF measurements. Histograms show FRET efficiencies determined for the 12-subunit RNAP (RNAP wt). The histogram was fitted with a double Gaussian function and the mean FRET efficiencies, E, the percentage distribution of the populations, A, and the coefficient of determination, R2, are given with SEs in the histograms. (E) Exemplary transients of an immobilized RNA polymerase show stable donor and acceptor fluorescence with no indication of multiple switching FRET states over several seconds and the resulting FRET transient until bleaching of the acceptor occurs (25 ms). Fluorescence transients were recorded using a TIRF microscopy setup with alternating laser excitation.