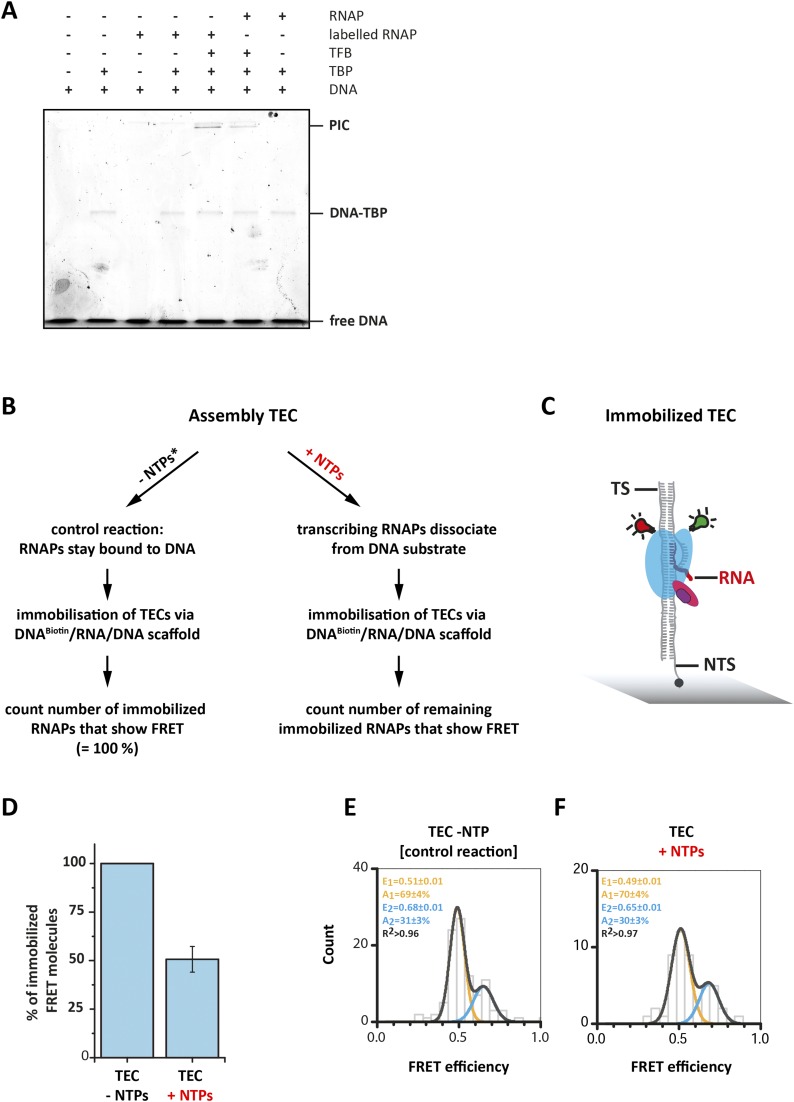
Fig. S1.
Modification of the RNAP does not interfere with preinitiation complex formation and transcriptional activity. (A) PIC formation was followed using an Alexa488-labeled SSV-T6 promoter DNA. Only if all components of the PIC are added is a slowly migrating fluorescent band corresponding to the PIC visible (0.4 µM RNAP, 8.7 µM TBP, 13 µM TFB, 133 nM DNA). The doubly labeled RNAP forms the PIC equally well as the unlabeled RNAP, indicating that the modified RNAP responds faithfully to the transcription initiation factors. (B) The transcriptional activity of donor/acceptor-labeled RNAPs. Double-labeled RNAPs were recruited to the elongation scaffold resulting in a TEC. Addition of NTPs and incubation of the sample at 65 °C starts transcription that ultimately leads to the dissociation of the RNAP when it reaches the end of the DNA substrate. Heparin was used as a scavenger that binds dissociated RNAPs, preventing reassociation of the released RNAP with the DNA substrate. These RNAPs will not be immobilized via the biotinylated elongation scaffold (biotin was positioned at the 5′-end of the NTS). (C) As a control, equal amounts of buffer instead of the NTP solution were added to the TEC, and all steps were performed as for the TEC+NTP sample. (D) The number of double-labeled RNAP molecules in the control (−NTP) vs. +NTP sample that are immobilized via the biotinylated elongation scaffold provides a direct readout about the number of double labeled RNAPs that were transcriptional active. NTP addition to the TEC led to a 51% reduction in the number of immobilized double-labeled RNAP molecules compared with the control sample providing evidence that the modified RNAPs are transcriptionally active. Other rare causes like spontaneous dissociation of the RNAP might occur but are not likely to contribute significantly to the total number of dissociated RNAPs. However, in this assay we are not able to differentiate between these processes. The FRET efficiency histograms derived for the control (E) and NTP incubated sample (F) are highly comparable exhibiting two FRET populations with the same relative amount of molecules in the corresponding populations. This demonstrated clearly that the transcriptionally active RNAPs are not depopulating a single “active” population but both RNAP populations are transcriptionally active. Data accumulated from three independent experiments.