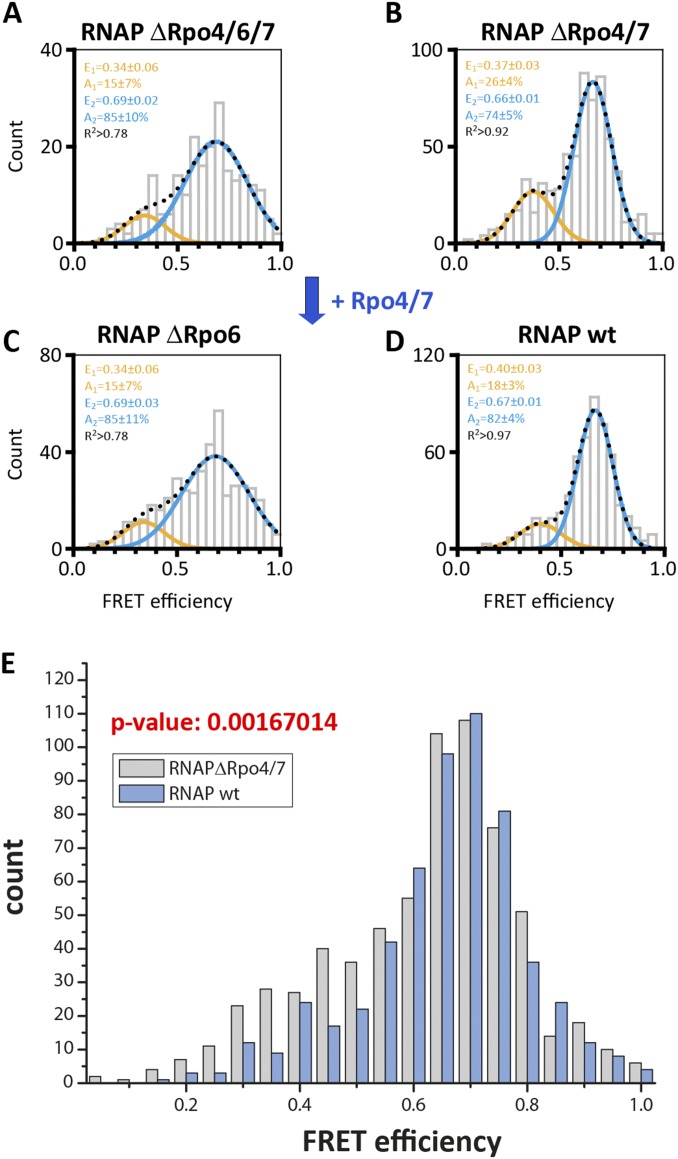
Fig. S3.
Conformation of the RNA polymerase clamp in the absence of subunit Rpo6 and Rpo4/7. Our recombinant archaeal model system allowed us to perturb the subunit composition of the labeled RNAPs. We prepared RNAP variants without the stalk subunits Rpo4/7 (RNAPΔRpo4/7) and without subunit Rpo6 that serves as docking site for Rpo4/7. Histograms showing the single-molecule FRET efficiencies determined for the 9-subunit RNAP (RNAP ΔRpo4/6/7; A) and 11-subunit RNAP (RNAP ΔRpo6; C) in comparison with the 10- and 12-subunit RNAP (RNAP ΔRpo4/7 and RNAP wt; B and D). RNAPs were immobilized via a biotin in subunit Rpo11 on a quartz slide for TIRF measurements. The histograms were fitted with a double Gaussian function and the mean FRET efficiencies, E, the percentage distribution of the populations, A, and the coefficient of determination, R2, are given with SEs in the histograms. Although addition of subunits Rpo4/7 to a RNAP ΔRpo4/7 enzyme slightly decreases the amount of low FRET population (E = 0.37), no shift was observed in an RNAP ΔRpo6 enzyme. Rpo6 is the docking site for subunits Rpo4/7, and hence, in its absence, Rpo4/7 cannot associate with the RNAP core. The absence of subunit Rpo6 leads to a very broad distribution, suggesting that Rpo6 might play a role in adjusting the position of the clamp. (E) Comparison of free RNAP wt vs. RNAP ΔRpo4/7 datasets: the null hypothesis that the datasets have the same distribution is rejected at the 5% level based on the Pearson χ2 test (P = 0.0016704 = 0.17%). In contrast, the null hypothesis that the datasets have the same distribution is not rejected when comparing and analyzing two RNAP wt or RNAP ΔRpo4/7 datasets. The Pearson χ2 test for RNAP wt datasets yielded P = 0.134966 (13.5%), and the Pearson χ2 test for RNAP ΔRpo4/7 datasets yielded P = 0.679731 (67.9%).