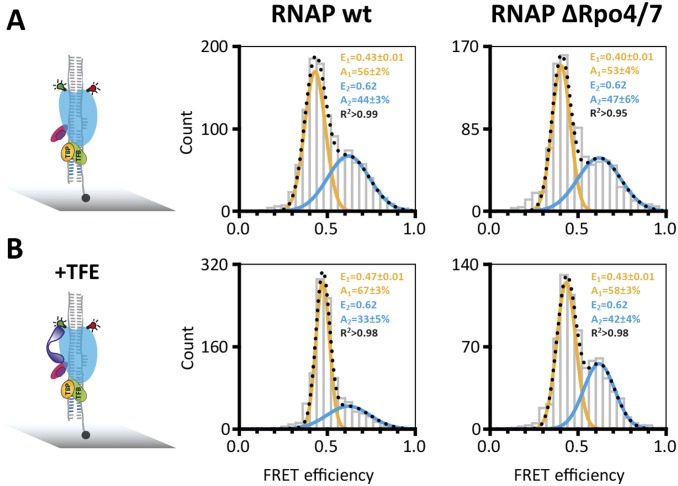
Fig. S4.
Conformation of the RNA polymerase clamp in transcription initiation complex. Histograms showing the single-molecule FRET efficiencies determined for the PIC in the absence (A) and presence (B) of TFE (purple). PICs were assembled using transcription initiation factors TBP (orange); TFB (green), the heterdouplex SSV T6 promoter DNA and labeled RNAP wt. PICs were immobilized via a biotinylated template strand on a quartz slide (gray) for TIRF measurements. On addition of TFE, an increased number of RNAP molecules are found in the low FRET state (+11%) indicative of a TFE-induced opening of the RNAP clamp. RNA polymerases that lack subunits Rpo4/7 do not respond to TFE. Subunits Rpo4/7 are known to be important for TFE binding, and the RNAPΔRpo4/7 variant serves as control for the authenticity of the TFE effect. The datasets were fitted with a double Gaussian function, and the mean FRET efficiencies, E, the percentage distribution of the populations, A, and the coefficient of determination, R2, are given with SEs in the histograms.