Figure 1. γKlotho is up-regulated in human triple negative breast cancer.
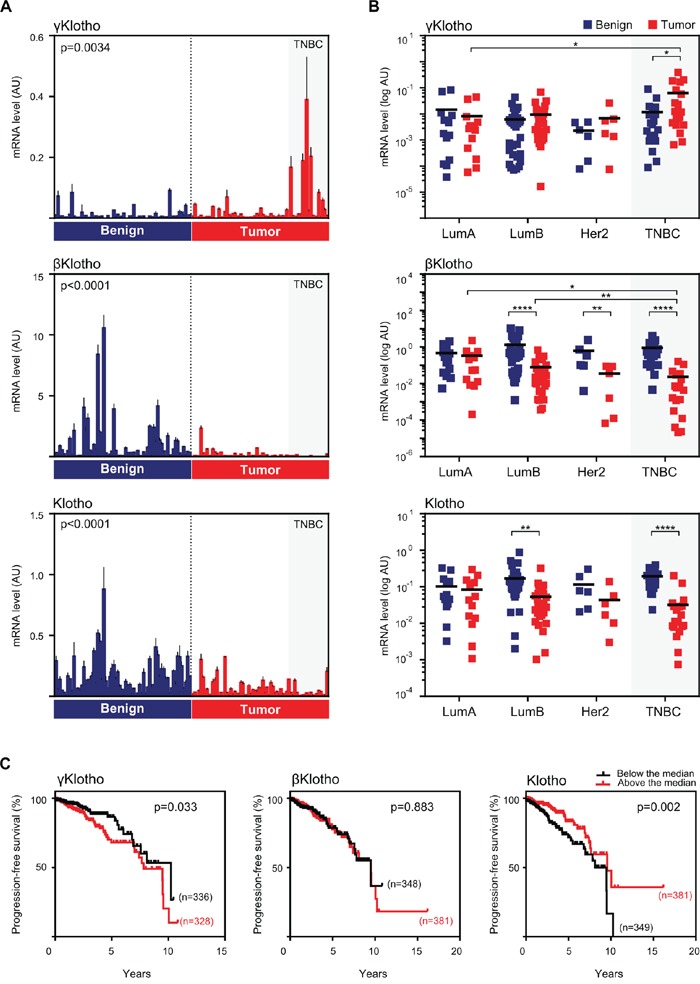
A. The expression of Klotho genes in normal/benign breast tissue (blue bars) and breast cancer (red bars). 68 samples of breast cancer specimens with corresponding patient-matched normal breast tissue were analyzed for mRNA expression of γKlotho, βKlotho, and Klotho by qRT-PCR. Expression levels were normalized against cyclophilin B. Each bar represents the mean ± SD of three replicates. Statistical analysis was performed on log-transformed data. Tumor and control groups were compared by paired t-test. p < 0.05 was considered statistically significant. B. In triple negative breast cancers γKlotho shows the opposite gene expression pattern than βKlotho and Klotho. Paired (benign and tumor) samples were divided into four groups according to the molecular subtype: luminal A (LumA; n = 13), luminal B (LumB; n = 30), HER2 (Her2; n = 6) and triple-negative breast cancer (TNBC; n = 19). The difference in gene expression between the subgroups was tested for significance using a two-way ANOVA followed by Bonferroni post-hoc tests on log-transformed data. Individual mRNA levels are presented on scatter dot plots using logarithmic scale for the y-axis. Black line denotes the mean. *p ≤ 0.05, **p.< 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. C. Kaplan-Meier progression-free survival curves according to the expression level with respect to the median of each Klotho gene in patients with invasive breast carcinoma with available triple negative status. Clinical and gene expression data were obtained from the TCGA portal. Log-rank (Mantel-Cox) tests were used to compare groups. Censored subjects are indicated on the curves by tick marks.
