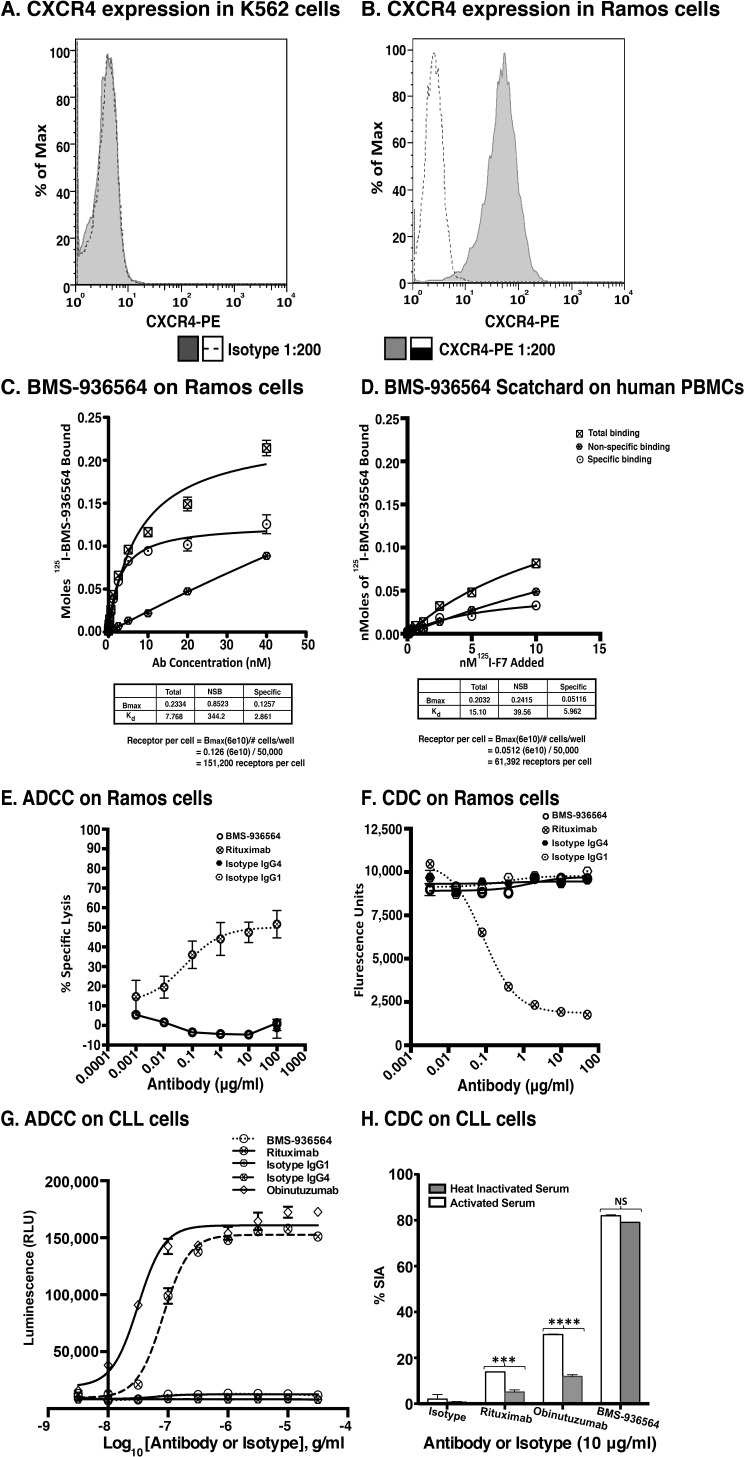
Figure 2. Scatchard analysis of Ulocuplumab (BMS-936564) binding to Ramos cells, human PBMCs and ADCC & CDC activity in Ramos cell line (Burkitt's lymphoma).
A. CXCR4 Expression profiling was done using an anti-CXCR4 antibody for surface staining in K562 and B. Ramos cell lines followed by analysis of samples using flow cytometry. The CXCR4 expression is presented in form of ΔMFI. C. The affinity of Ulocuplumab (BMS-936564) was measured using endogenously expressed CXCR4 positive Ramos cells. The binding capacity of 125I-Ulocuplumab (BMS-936564) to CXCR4+ cells in presence or absence of excess unlabeled antibody was determined and a Kd of 2.9 nM and approximately 1.5×105 receptors/cell was calculated. D. Normal peripheral blood cells were incubated with increasing concentrations of 125I-Ulocuplumab (BMS-936564). The counts per minute bound (CPM) in the presence or absence of unlabeled antibody was measured. Scatchard analysis of saturation binding curves was performed with Prism 4 GraphPad software (GraphPad software, San Diego, CA) using nonlinear regression analysis. The symbol  ,
,  and
and  represent total binding (TB), non-saturable binding (NSB), and saturable binding (SB), respectively. E. Ramos cells (target) were labeled with bis(acetoxymethyl) 2,2′:6′,2″-terpyridine-6,6″-dicarboxylate (BADTA). Freshly isolated human peripheral blood mononuclear cells PBMCs (effector) were used for allotyping of the Ulocuplumab (BMS-936564). Human PBMCs were cultured with labeled Ramos cells at 1:50 ratio of target/effector cell (T/E) ratios in the presence or absence of rituximab, Ulocuplumab, or isotype (BMS-936564) for 1 h at 37°C. Ramos cells alone served as spontaneous release (SR) and Ramos cells lysed with 1% Triton X-100 served as total release (TR). The lysis was measured by using europium (Eu)-based detection. BMS-936564, Rituximab and their respective isotypes with varied concentrations were tested using Ramos cell line. F. Cell-based CDC assay of Ulocuplumab (BMS-936564) /Rituximab and their respective isotypes as controls: lysis of Ramos cells in the presence of human complement was measured by using Alamar Blue release. G. The stable transfected Jurkat cell line expressing FcgRIIIa and NFAT-RE luc was used as effector E. in an ADCC Reporter Bioassay from Promega. CLL cells (target, T) were plated in ratio of 1:1 with the effector cells. The effector: target cells were incubated in the presence or absence of rituximab or Ulocuplumab (BMS-936564), or obinutuzumab between 0.001-30 ug/ml of concentrations for 6 hrs at 37°C. Rituximab and obinutuzumab were used as positive controls. The reaction was developed by incubating the cells with Bio-GloTM reagent for 30 minutes at room temperature in dark. The plates were read on luminometer and following the background subtraction, relative-light units (RLU) were calculated for different antibodies/isotypes using GraphPad Prism software. H. Complement dependent cytotoxicity (CDC) for 10 μg/ml of either Ulocuplumab (BMS-936564), rituximab, obinutuzumab or isotype was tested in CLL cells after incubation with either 5% fresh human or heat inactivated serum (to denature complement) was measured by using CD19/CD5/Annexin V staining followed by flow cytomerty analysis. Rituximab and obinutuzumab were used as positive controls. The data are the mean and SD of triplicate cultures. The statistical data was analyzed using Bonferroni correction test in GraphPad Prism software.
represent total binding (TB), non-saturable binding (NSB), and saturable binding (SB), respectively. E. Ramos cells (target) were labeled with bis(acetoxymethyl) 2,2′:6′,2″-terpyridine-6,6″-dicarboxylate (BADTA). Freshly isolated human peripheral blood mononuclear cells PBMCs (effector) were used for allotyping of the Ulocuplumab (BMS-936564). Human PBMCs were cultured with labeled Ramos cells at 1:50 ratio of target/effector cell (T/E) ratios in the presence or absence of rituximab, Ulocuplumab, or isotype (BMS-936564) for 1 h at 37°C. Ramos cells alone served as spontaneous release (SR) and Ramos cells lysed with 1% Triton X-100 served as total release (TR). The lysis was measured by using europium (Eu)-based detection. BMS-936564, Rituximab and their respective isotypes with varied concentrations were tested using Ramos cell line. F. Cell-based CDC assay of Ulocuplumab (BMS-936564) /Rituximab and their respective isotypes as controls: lysis of Ramos cells in the presence of human complement was measured by using Alamar Blue release. G. The stable transfected Jurkat cell line expressing FcgRIIIa and NFAT-RE luc was used as effector E. in an ADCC Reporter Bioassay from Promega. CLL cells (target, T) were plated in ratio of 1:1 with the effector cells. The effector: target cells were incubated in the presence or absence of rituximab or Ulocuplumab (BMS-936564), or obinutuzumab between 0.001-30 ug/ml of concentrations for 6 hrs at 37°C. Rituximab and obinutuzumab were used as positive controls. The reaction was developed by incubating the cells with Bio-GloTM reagent for 30 minutes at room temperature in dark. The plates were read on luminometer and following the background subtraction, relative-light units (RLU) were calculated for different antibodies/isotypes using GraphPad Prism software. H. Complement dependent cytotoxicity (CDC) for 10 μg/ml of either Ulocuplumab (BMS-936564), rituximab, obinutuzumab or isotype was tested in CLL cells after incubation with either 5% fresh human or heat inactivated serum (to denature complement) was measured by using CD19/CD5/Annexin V staining followed by flow cytomerty analysis. Rituximab and obinutuzumab were used as positive controls. The data are the mean and SD of triplicate cultures. The statistical data was analyzed using Bonferroni correction test in GraphPad Prism software.