Figure 3. Identification of FAT4 as a tumor suppressor gene in HCC.
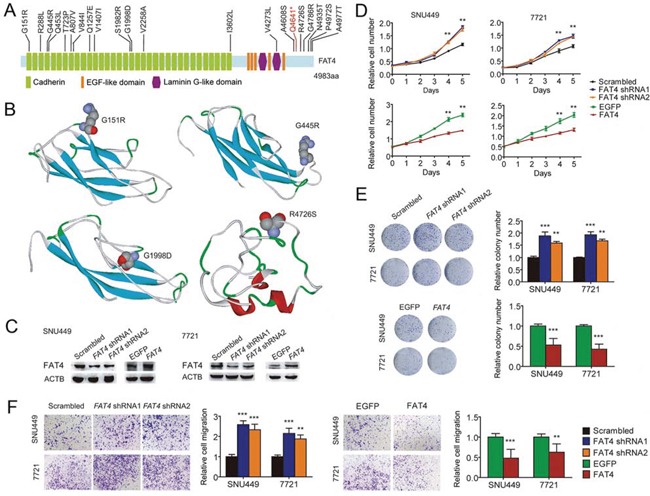
A. Schematics of protein alterations in FAT4 caused by somatic mutations. B. Structural modeling showing locations of the mutations G151R, G445R, G1998D, and R4672S. C. Protein blots showing FAT4 knockdown with shRNA and overexpression with TALE in indicated cells, compared with their respective controls. D. Growth curves showing accelerated growth with FAT4 knockdown and decelerated growth with FAT4 overexpression in indicated cells. E. Colony formation showing increased clones with FAT4 knockdown and decreased clones with FAT4 overexpression in indicated cells. F. Cell migration showing elevated migration with FAT4 knockdown and reduced migration with FAT4 overexpression in indicated cells. Experiments were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001.
