Figure 5. The proposed binding region of the REIC/DKK-3-SGTA interaction.
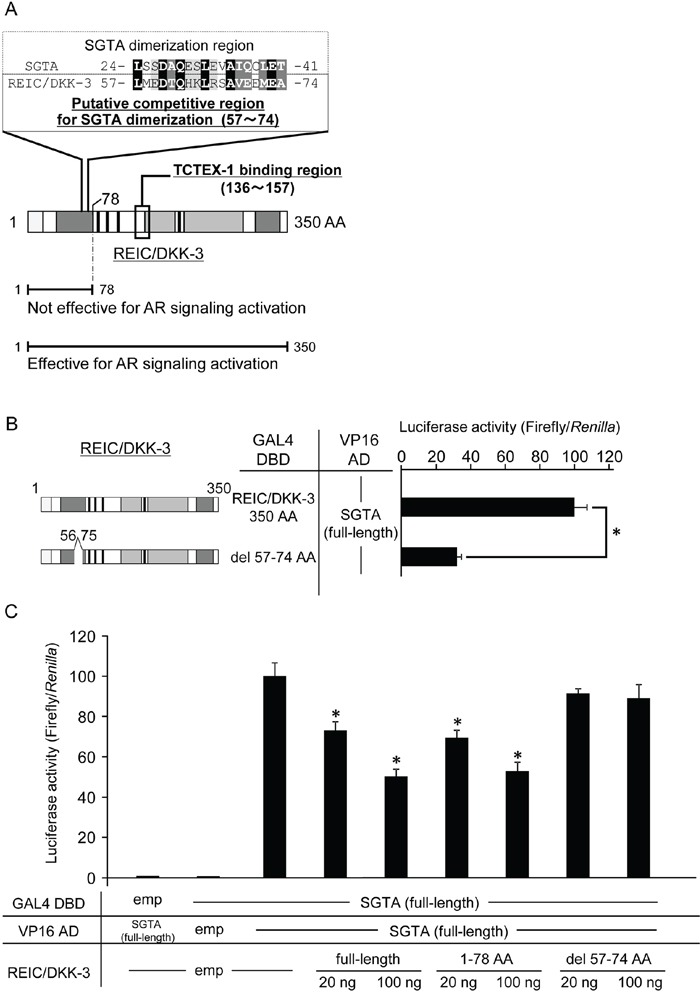
A. Based on the results from Figure 2, sequence homology analysis was performed between the 1–78 AA region of REIC/DKK-3 (SGTA binding region) and the reported SGTA dimerization region (1–80 AA) [24,30,31]. The analysis revealed the putative competitive region (57–74 AA) of REIC/DKK-3 that affects SGTA dimerization. Sequence alignment analysis between the competitive region of REIC/DKK-3 and the affected part (24–41 AA) of the SGTA dimerization region was performed using the ClustalW software program (http://www.genome.jp/tools/clustalw/). The completely conserved residue is shaded in black, and the residues belonging to strongly similar groups are shaded in gray. The residues exhibiting weaker similarity are shaded in light gray. B. The interactions of REIC/DKK-3, which was deleted the putative binding region (57–74 AA), with SGTA and full-length SGTA was examined in a mammalian two-hybrid assay. The left panel depicts the REIC/DKK-3 fragment examined, and the results are shown in the right panel. The data are from three independent experiments. The bar graphs show the luciferase activities on the top column as 100%. *, A significant difference was observed. C. The interference activity of REIC/DKK-3 (del 57–74) with the SGTA-SGTA dimerization was analyzed in a mammalian two-hybrid assay. The data are from three independent experiments. The bar graphs show the luciferase activities on the interaction of SGTA-DBD, SGTA-VP16 AD and HA-tagged empty vector column as 100%. *, A significant difference was observed.
