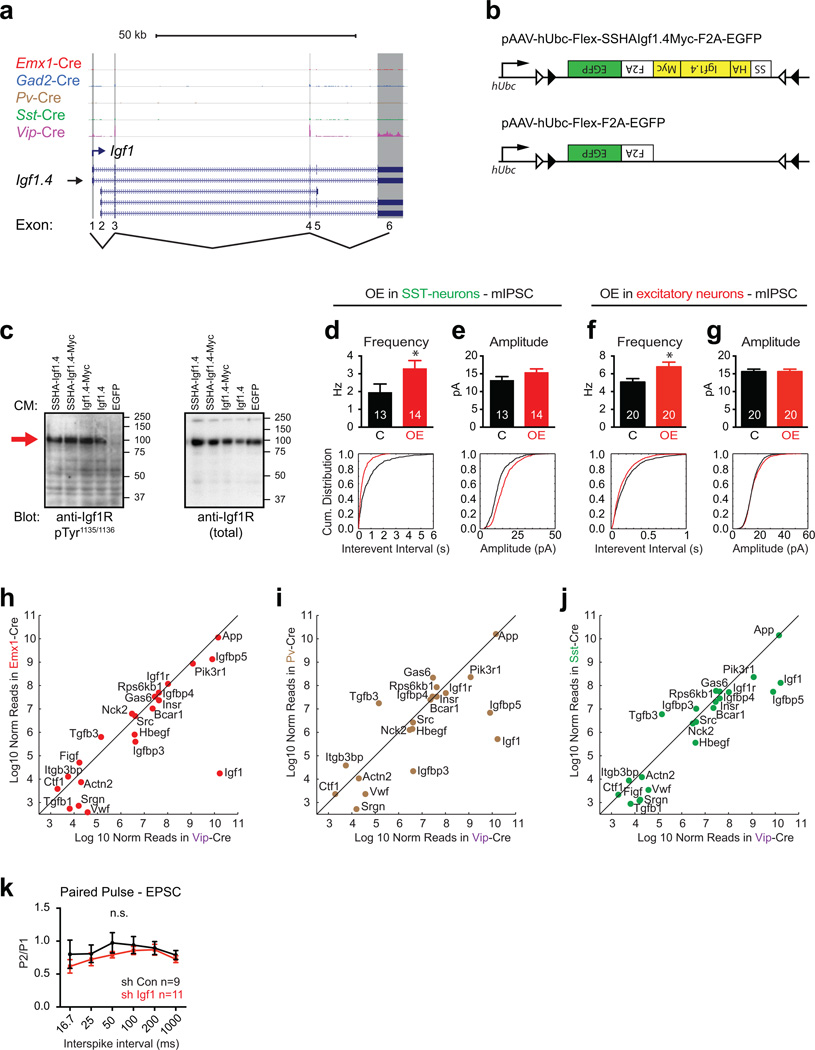
Extended Data Figure 7. Effects of IGF-1 overexpression in excitatory and Sst-positive neurons.
a) RiboTag-Seq identifies Igf1.4 as the major Igf1-isoform expressed in VIP neurons. Representative tracks of histograms of the RNA-Seq reads in each Cre line across the Igf1 genomic locus. Data is from the 7.5 hour light exposure RiboTag-Seq datasets.
b) AAV constructs for the Cre-dependent expression of HA-/Myc-tagged IGF-1 (Igf1.4) and EGFP (i.e. IGF1-OE, top) or of EGFP alone (i.e. control, bottom). F2A = Furin cleavage site followed by the 2A peptide; black and white triangles represent a Cre-dependent Flex-switch.
c) Western blot analysis of IGF1-receptor activation status in lysates of serum starved HEK293T cells that were stimulated with conditioned media (CM) containing epitope-tagged isoforms of IGF-1. CM was produced by transfecting HEK293T cells with the respective construct and collecting the culture media. IGF1-receptor is detected with antibodies against either activated IGF1-receptor (anti Igf1R pTyr1136/1138) or total IGF-1 receptor (anti Igf1R total). (Molecular weight markers are on the right and the arrow indicates the band of the IGF1-receptor).
d,e) Bar graphs and cumulative distribution plots showing mIPSC frequency / interevent interval (d) and amplitude (e) of mIPSCs recorded from EGFP positive neurons in P20 Sst-Cre mice that were intracortically injected with AAVs driving the expression of control (C) or IGF1-OE (OE) constructs (Amplitude, p = 0.16; Frequency, p = 0.01; Mann Whitney U-Test; numbers inside bars indicate the number of cells recorded).
f,g) Bar graphs and cumulative distribution plots showing mIPSC frequency / interevent interval (f) and amplitude (g) of mIPSCs recorded from EGFP positive neurons in P20 Emx1-Cre mice that were intracortically injected with AAVs driving the expression of control (black, n = 20) or IGF1-OE AAVs (red, n = 20). (Amplitude, p = 0.99; Frequency, p = 0.01, Mann Whitney U-Test)
h–j) Scatter plots of IGF-1-interacting proteins showing the Log10 normalized mean expression values in Vip-Cre neurons versus each of the other Cre lines (h - Emx1, i - Pv, j - Sst).
k) Quantification of EPSC paired-pulse recordings from VIP neurons infected with control shRNA (black n = 9) or Igf1 shRNA (red n = 11) expressing AAVs. The ratio of the second EPSC amplitude divided by the first EPSC amplitude is plotted against inter-stimulus interval (p = 0.1, two-way ANOVA).