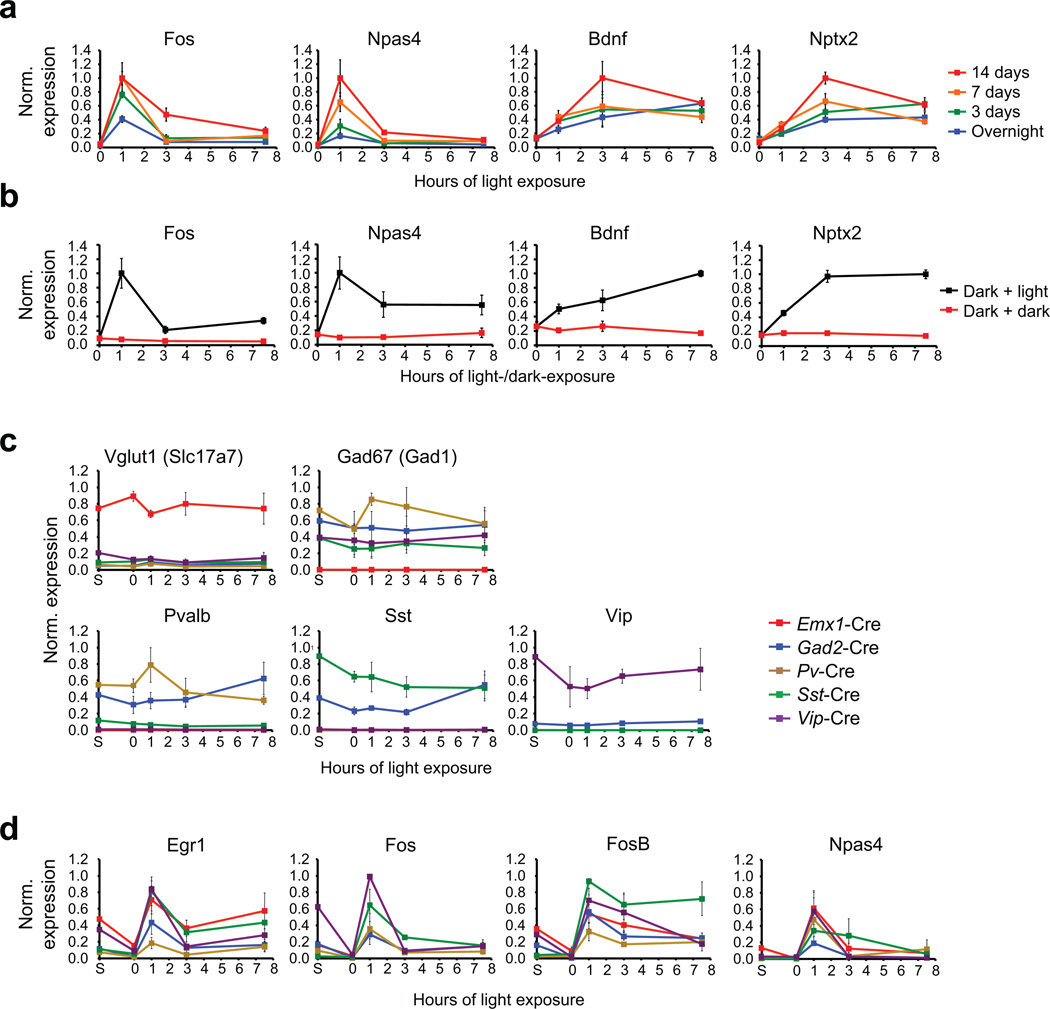
Extended Data Figure 1. Validation of the sensory stimulation protocol and the RiboTag-based cell-type-specific purification of mRNA.
a) Quantitative real-time PCR (qPCR) for known experience-regulated genes on RNA isolated from the visual cortex of mice that were dark-housed for varying durations (overnight, 3 days, 7 days or 14 days) and then either sacrificed in the dark or exposed to light for 1, 3, or 7.5 hours, and then sacrificed. Data are normalized to the maximal value in each dataset and represent the mean and standard error of four biological replicates.
b) qPCR for known experience-regulated genes on RNA isolated from the visual cortex of mice that were dark-housed for 14 days and then either exposed to light for 1, 3, or 7.5 hours (dark + light, black) or kept in the dark during these hours (dark + dark, red). All mice of a given time point were dissected in very close temporal proximity. Data are normalized to the maximal value in each dataset and represent the mean and standard error of four biological replicates.
c) qPCR for known cell-type-specific marker genes on RNA isolated from RiboTag mice expressing Cre in distinct neuronal subtypes. Data are normalized to the maximal value in each dataset and represent the mean and standard error of three biological replicates.
d) qPCR for known early-induced transcription factors on RNA isolated from RiboTag mice expressing Cre in distinct neuronal subtypes. Data are normalized to the maximal value in each dataset and represent the mean and standard error of three biological replicates.