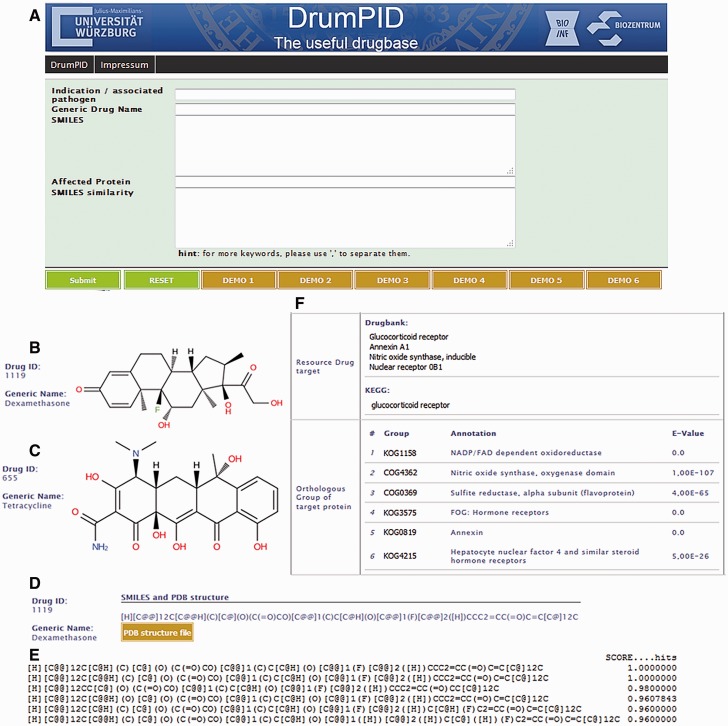
Figure 2.
DrumPID search capabilities. DrumPID allows the user to explore potential antibiotic lead structures, optimizing predictions from animal tests or explore the chemical space around a compound together with the affected protein interaction networks. For each capability, DrumPID makes direct calculations based on the chemical properties of the drug as well as collating and comparing information from several source of databases (database logic rules show all original database sources available) and its own stored data (see text for details). (A) Web interface. DrumPID allows to search for Indications and associated Pathogens, generic drug names, SMILES, drug-affected proteins as well as similar substructure of SMILES. (B) Drug indication query (hematological disorder). Example: the drug Dexamethasone with corresponding structure and Drug ID, scroll down for more information (not shown). (C) Pathogen query. Example: drug Tetracycline (structure) against Borrelia burgdorferi (B. burgdorferi). There is further information on treatment, drug usage as well as chemical and biological properties (not shown). (D) SMILES search. Example: [H][C@@]12C[ C@@H](C)[C@](O)(C (=O)CO)[C@@]1( C)C[C@H](O)[C@ @]1(F)[C@@]2([H])CCC2 = CC(= O)C = C [C@]12C. The resulting drug Dexamethasone is shown. Furthermore, SMILES notation is converted into PDB structure files, which enables further studies of the compounds, e.g. docking studies. (E) SMILES similarity search. In addition, to identify drugs consisting similar substructures, a similarity search for SMILES is possible (Tanimoto similarity score > 0.66). For example, using the SMILES [H][C@@] 12C[C@@H] (C)[C@](O)(C( =O)CO) [C@@]1(C)C[C@H] (O)[C@@]1(F)[C@@]2([H])CCC2 = CC(=O)C = C[C@]12C calculates Dexamethasone and Betamethasone with a similarity score of 1 as top hits (here hits >0.96 are shown). (F) Protein interactions. For each drug, known targets and pathways are given (including source scheme; here only targets shown). For all targets there is Ortholog group search (COG/KOG) including annotations and E-values. Furthermore, output entries carry links including other interaction databases (PlateletWeb, AnDom, GoSynthetic, HPRD, iHop, STRING and KEGG) are available (not shown). Example: Glucocorticoid receptor gave 37 results, four Protein interactions and six Ortholog Groups for the drug Dexamethasone. (For more details, see text and tutorials in supplementary material.)