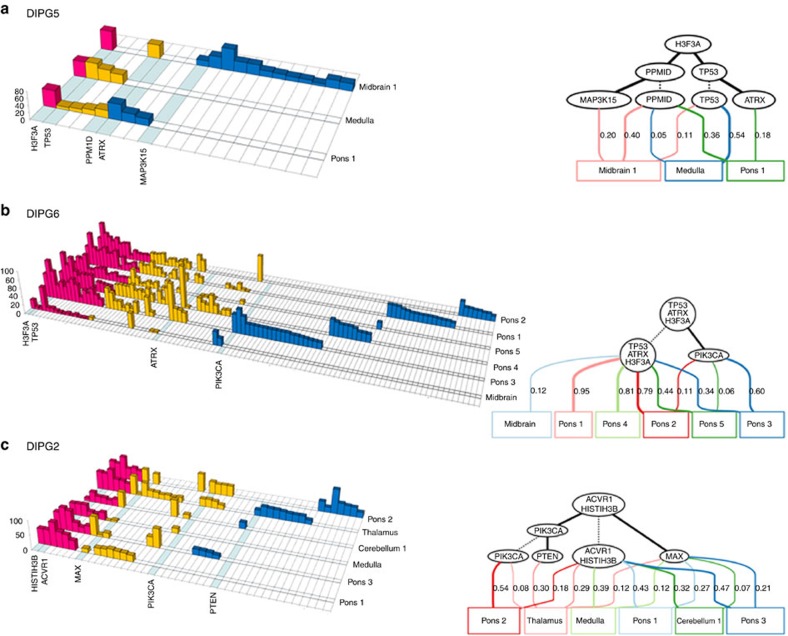
Figure 2. Selected examples of clonal evolution within DIPG tumours.
Left: histograms show the raw allele frequencies (whole exome sequencing data) for each somatic mutation in different autopsy regions within each tumour. Red: ubiquitous mutations across regions; yellow: mutations shared in at least two regions; blue: mutations seen in only one region. Right: phylogenetic trees constructed from the mutation allele frequencies of deep amplicon sequencing data showing the order of evolution along with support probabilities (upper portions of graphs) and clonal mixing proportions within samples (lower portions). For clarity, only mutations selected to be likely oncogenic are shown. (a) DIPG5: a rare case harboring both TP53 and PPM1D mutations, which are generally found to be mutually exclusive. PPM1D and TP53 mutations occur in distinct clones and are both secondary to H3K27M. ATRX is also secondary and subclonal. (b) DIPG6: while it is impossible to resolve the order of H3/TP53/ATRX mutations' appearance, PIK3CA is clearly sub-clonal and appears in the later stages of evolution within this tumour. (c) DIPG2: the H3.1 K27M and ACVR1 main driver mutations are ubiquitous, occur at similar frequencies across all samples, and their mutation order cannot be resolved. Conversely, other accessory driver mutations are clearly secondary in order of appearance, and are present only in distinct subclones.