Figure 4.
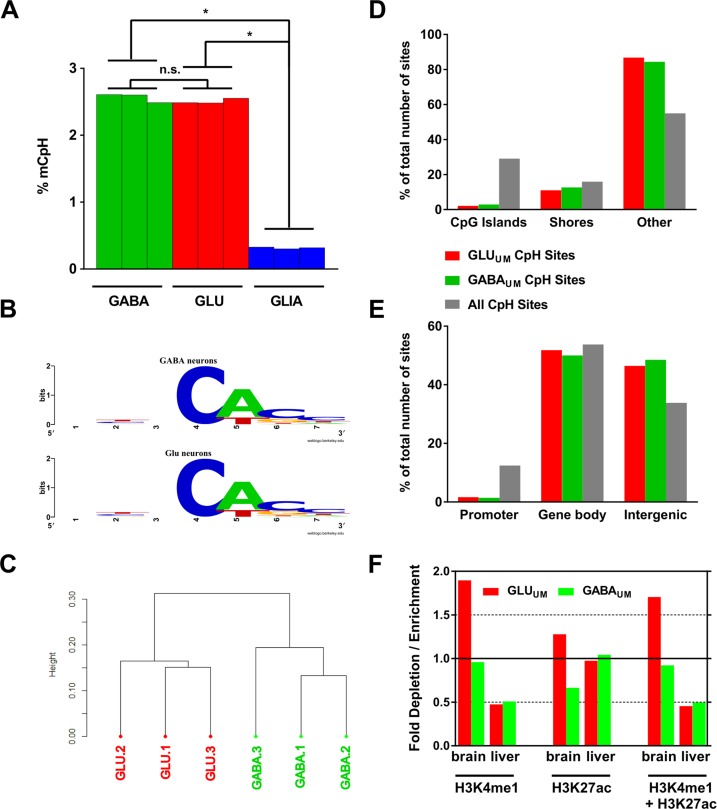
Analysis of non-CpG (CpH) methylation measured by ERRBS. (A) Average CpH methylation in GABA (green) and GLU (red) neurons, and in GLIA (blue); N = 3 samples for each cell type. Data for GLIA are from (32); *P-values < 0.0001 for comparisons of GLIA with GABA or GLU (unpaired t-test). (B) Sequence context of CpH methylation in GABA and GLU neurons. In both neuronal subtypes CpH methylation occurs in mostly the same context—CACC. Methylated CpH cytosine is shown at position 4. (C) Unsupervised hierarchical clustering of CpH methylation data for GLU and GABA samples. 1, 2, 3 denote individual subjects. (D) Enrichment / depletion of CpH DM sites within CpG island-related genomic features. The data presented as% of total number of sites within each group (GLUUM, GABAUM and all CpH sites present in the ERRBS data set). (E) Enrichment/depletion of CpH DM sites within promoters, gene bodies and intergenic regions. (F) Enrichment of the distal CpH DM sites within predicted enhancers defined based on histone modification profiling data from REMC (74) (see Figure 3F for details).