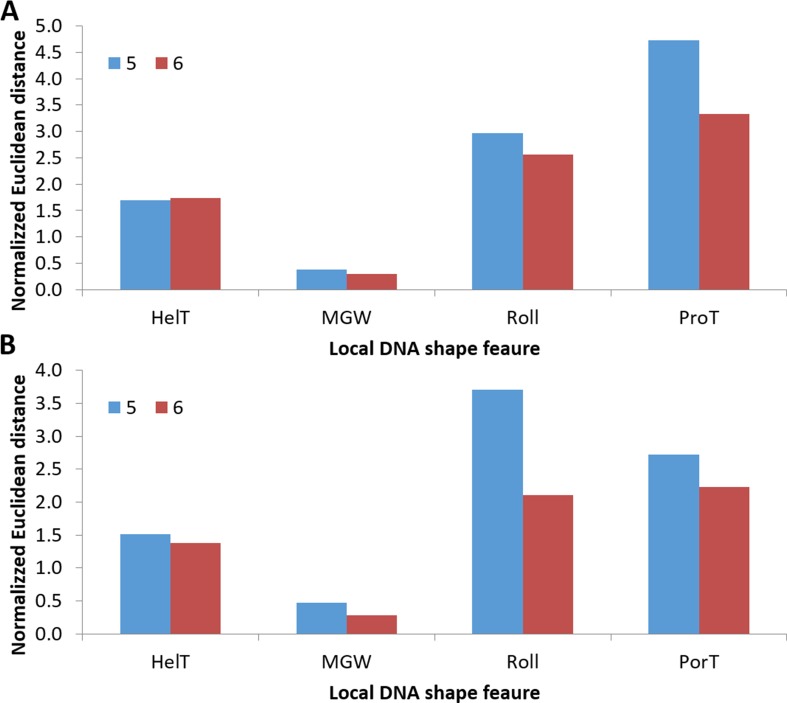
Figure 7.
Similarity of QPID ATF1 binding preferences to PBM and ChIP-seq. Normalized Euclidean distance between DNA local shape feature predictions of oligonucleotide 5 and 6 and average shape feature predictions of CRE half-sites binding sites (TGACG) were calculated (see Materials and Methods). (A) Distance to an average of 1647 BSs measured by PBM. (B) Distance to an average of 4310 BSs measured by ChIP-seq. HelT, MGW, Roll and ProT: helix twist, minor groove width, roll and propeller twist.