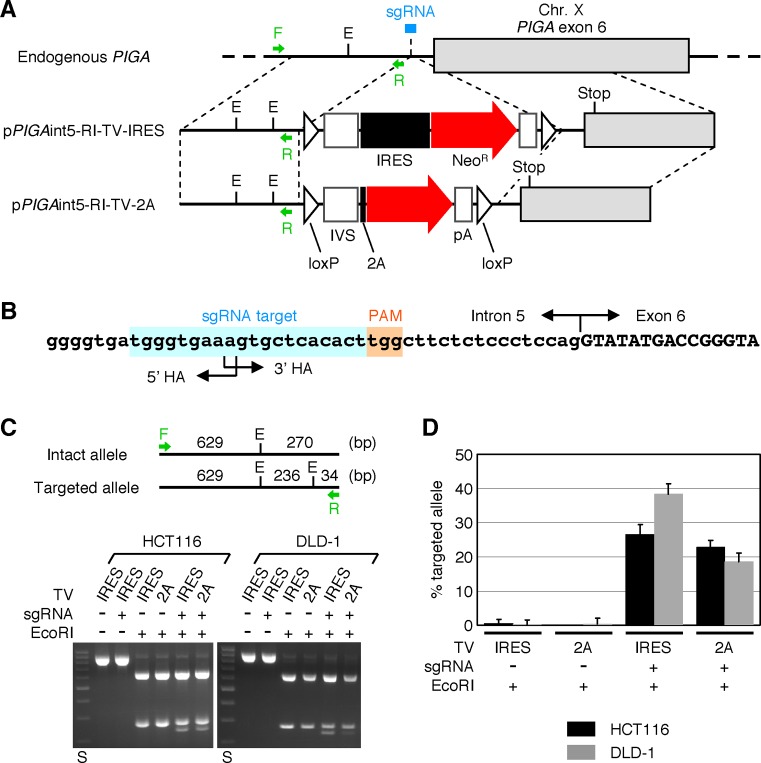
Figure 6.
H/R ratios of plasmid-based targeting vectors in CRISPR-Cas9-assisted gene targeting. (A) A scheme describing the targeting of PIGA intron 5 with the assistance of a site-specific CRISPR-Cas9 endonuclease. Arrows associated with F or R indicate PCR primers used for RFLP analysis shown in (C). sgRNA: single guide RNA as a component of CRISPR-Cas9 system. E: EcoRI site. Other abbreviations are shown in the legends for Figures 1–3. (B) A sgRNA target sequence located at the junction of 5′ and 3′ homology arms. The genomic sequence of PIGA marked with a sgRNA target site, protospacer adjacent motif (PAM) and intron 5/exon 6 boundary is shown. (C) RFLP analysis. The HCT116 and DLD-1 cell lines were co-transfected with the above-described CRISPR-Cas9 cleavase along with either pPIGAint5-RI-TV-IRES (IRES) or pPIGAint5-RI-TV-2A (2A) and selected with G418. gDNA extracted from G418-resistant cells were processed for PCR amplification with the primer pair shown in (A), digested with EcoRI and separated by agarose gel electrophoresis. (Top) Deduced cleavage patterns of PCR products amplified from intact and targeted PIGA alleles. (Bottom) Representative gel images. A plasmid expressing Cas9 and a sgRNA without 20-nt guide RNA was used as a control for the CRISPR-Cas9 cleavase. DNA size standards (S) include 200–1000 bp DNA fragments with an interval of 100 bp. (D) The ratio of targeted versus entire PIGA alleles determined based on the intensities of 270 and 236-bp bands (mean ± s.e.m.; n = 3).