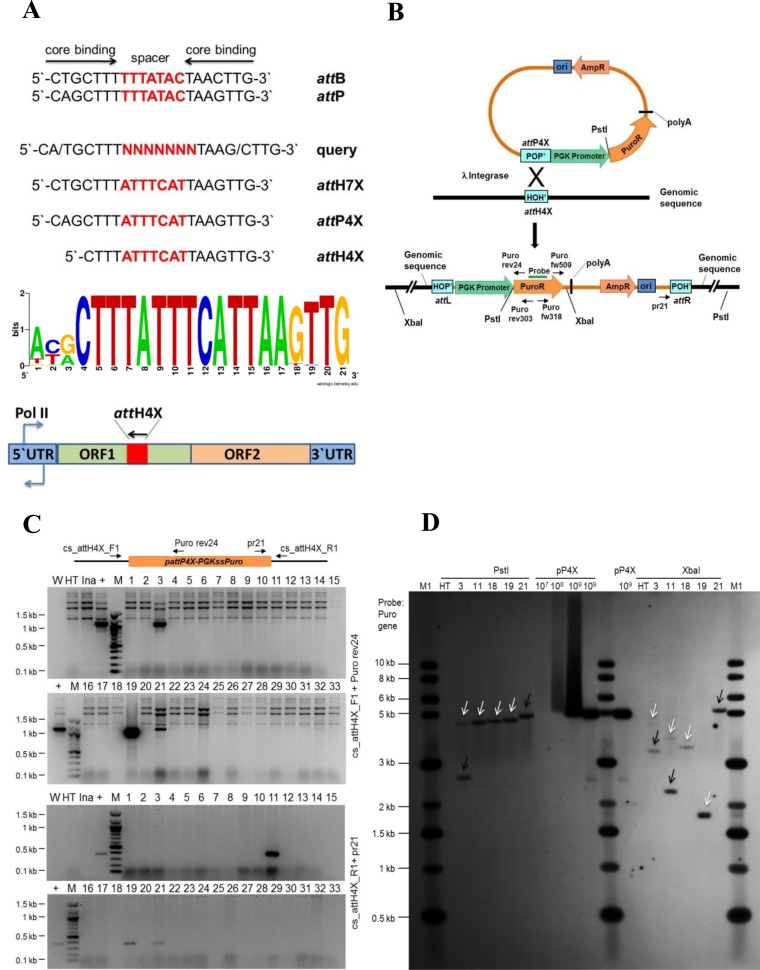
Figure 1.
Targeting endogenous attH4X in HT1080 cells. (A) Diagram showing the 21 nucleotide sequences comprising the core binding and spacer sequences of various att sites and features of the LINE-1 retrotransposon. Wild type attB and attP sequences aligned to show the respective core binding and spacer sequences. The query sequence was used in a bioinformatics search for targets resembling core att sites. Sequence logo analysis was performed for the 18 bp attH4X sequences in LINE-1 elements in the targeted cell lines including additional three nucleotides at the 5′ end in order to compare with the 21 bp attB sequence. A LINE-1 retrotransposon diagram with the position and orientation of attH4X is shown at the bottom. See text for details. (B) Schematic drawing showing pattP4X-PGKssPuro target vector and predicted recombination between attP4X and genomic attH4X. Positions of relevant primers (Puro rev24, Puro rev303, Puro fw318, Puro fw509 and pr21), restriction sites and the probe used for Southern blotting are indicated. (C) Screening for attH4X × attP4X recombination events in HT1080 clones. PCR was performed with genomic DNA and primers cs_attH4X_F1 and Puro rev24 (for attL junction) and cs_attH4X_R1 and pr21 (for attR junction). PCR amplified products of the expected size (1100 bp; for the left junction) were detected in clones 3, 19 and 21 (top two panels) and (∼375 bp; for the right junction) in clones 11, 19 and 21 (bottom two panels). W, no DNA template control; HT, negative control (genomic DNA from parental cells); Ina, genomic DNA from puromycin resistant clones obtained through co-transfection of pattP4X-PGKssPuro and pCMVssIna, the latter expressed inactive Int as negative control; +, positive control (genomic DNA from a HT1080 clone carrying an attH4X x attP4X integration event); M, 100 bp DNA ladder; 1 to 33, genomic DNA from puromycin resistant HT1080 clones obtained through co-transfection of pattP4X-PGKssPuro and pCMVssInt-C3CNLS. (D) Southern blot analysis. Genomic DNA purified from five targeted HT1080 clones, as indicated, and parental HT1080 cell line was subjected to digestion with PstI or XbaI. A PCR-derived digoxigenin-labeled probe complementary to the puromycin resistance gene was used. Lanes: M1, 1 kb DNA ladder; HT, genomic DNA from HT1080; 3, 11, 18*, 19 and 21, genomic DNA from targeted clones; pP4X (107, 108, 109), copies of linearized pattP4X-PGKssPuro loaded as positive control. The arrows indicate fragments of expected size for clones 3 and 11. *HT 1080 Clone 18 (carrying a single-copy transgene) was obtained from a screen of puromycin-resistant HT1080 clones that resulted from a previous co-transfection of pattP4X-PGKssPuro and pCMVssInt-h/218. White arrow heads indicate fragments of the expected size and black arrow heads indicate extra or unexpected fragments in the targeted clones.