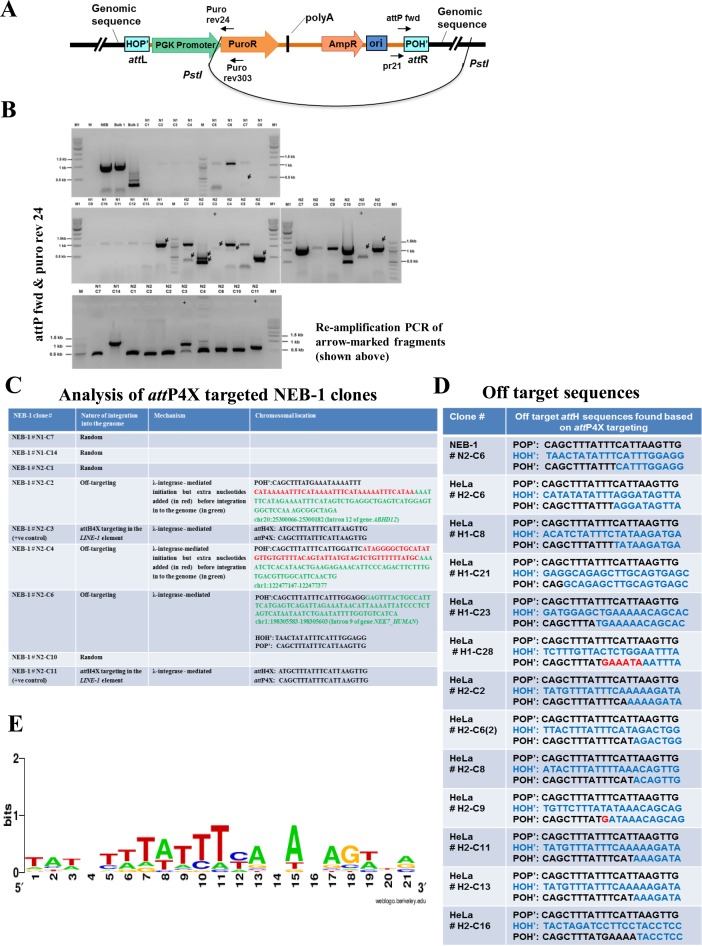
Figure 6.
Off-target analysis in NEB-1 and HeLa cell lines. (A) Schematic drawing showing self-ligation of PstI digested genomic DNA consisting of integrated pattP4X-PGKssPuro target vector. Positions of relevant primers (Puro rev24, Puro rev303, pr21, attP fwd) used for inverse nested PCR to determine the potential attR site are indicated. (B) Screening for off-targeting events in NEB-1 clones. Inverse nested PCR was performed using genomic DNA from 26 puromycin-resistant NEB-1 clones which were obtained from two independent transfections. PCR products that were not present in control PCRs (with genomic DNA of NEB-1 parental cells as template) and were larger than 200 bp (fragments marked with black arrows) were detected in clones N1-C7, N1-C14, N2-C1, N2-C2, N2-C3, N2-C4, N2-C6, N2-C10, N2-C11 (upper two panels). They were extracted and re-amplified (bottom panel) for sequencing. W, no DNA template control; NEB, negative control (genomic DNA from parental cells); Bulk 1 & bulk 2, genomic DNA template from all puromycin resistant colonies obtained through two independent co-transfections of pattP4X-PGKssPuro and pCMVssInt-h/218CNL; N1(C1-C14) and N2 (C1-C12), genomic DNA from puromycin resistant NEB-1 clones obtained through two independent co-transfections of pattP4X-PGKssPuro and pCMVssInt-C3CNLS; +, positive control (genomic DNA from a NEB-1 clone carrying an attH4X × attP4X integration event); M, 100 bp DNA ladder; M1, 1 kb DNA ladder. (C) Table showing sequence analysis of NEB-1 targeted clones (nature, possible mechanism and chromosomal location of genomic integration of target vector). (D) Table showing the observed off-target sequences based on attP4X targeting in NEB-1 and HeLa cell lines. (E) Sequence logo analysis for the 21bp off-target attH (HOH‘) sequences found based on attP4X targeting in NEB-1 and HeLa cell lines.