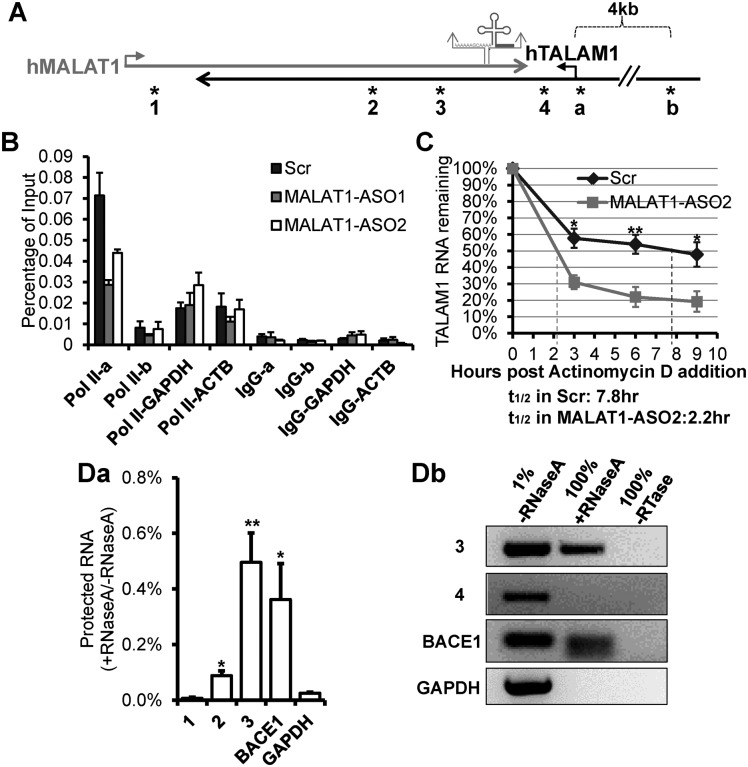
Figure 2.
MALAT1 positively regulates the transcription and stability of TALAM1, and forms RNA duplex with TALAM1 in vivo. (A) Schematic diagrams depicting MALAT1 genomic locus. Relative positions of the primers used in ChIP experiment in (B), are indicated by asterisks labeled as ‘a’ and ‘b’: primer set ‘a’ is at TALAM1 promoter, ‘b’ is at upstream 4 kb of TALAM1 TSS. Relative positions of the primers analyzed in RNase protection assay in (D), are indicated by asterisks labeled as 1 to 4. (B) ChIP-qPCR of RNA Pol II at TALAM1 promoter region, in HeLa cells treated with Control-Scr, MALAT1-ASO1 and ASO2. Primers from 4 kb upstream of TALAM1 TSS, GAPDH and ACTB promoters were used as negative controls. (C) TALAM1 stability in HeLa cells treated with Scr or MALAT1-ASO2. Dotted lines represent the half-life of TALAM1. (D) In vivo RNase protection assay. HeLa cells were lysed in absence or presence of RNaseA, then RNA was extracted and analyzed via (Da) RT-qPCR and (Db) RT-PCR, using primers illustrated in (A). Data in (B) and (C) are mean values with SEM, NB = 3 (3 technical replicates of representative biological replicates), NC = 4; Data in (Da) is mean value with SD, N = 3 (biological replicates, *P < 0.05, **P < 0.01, two-tailed paired Student t-test).