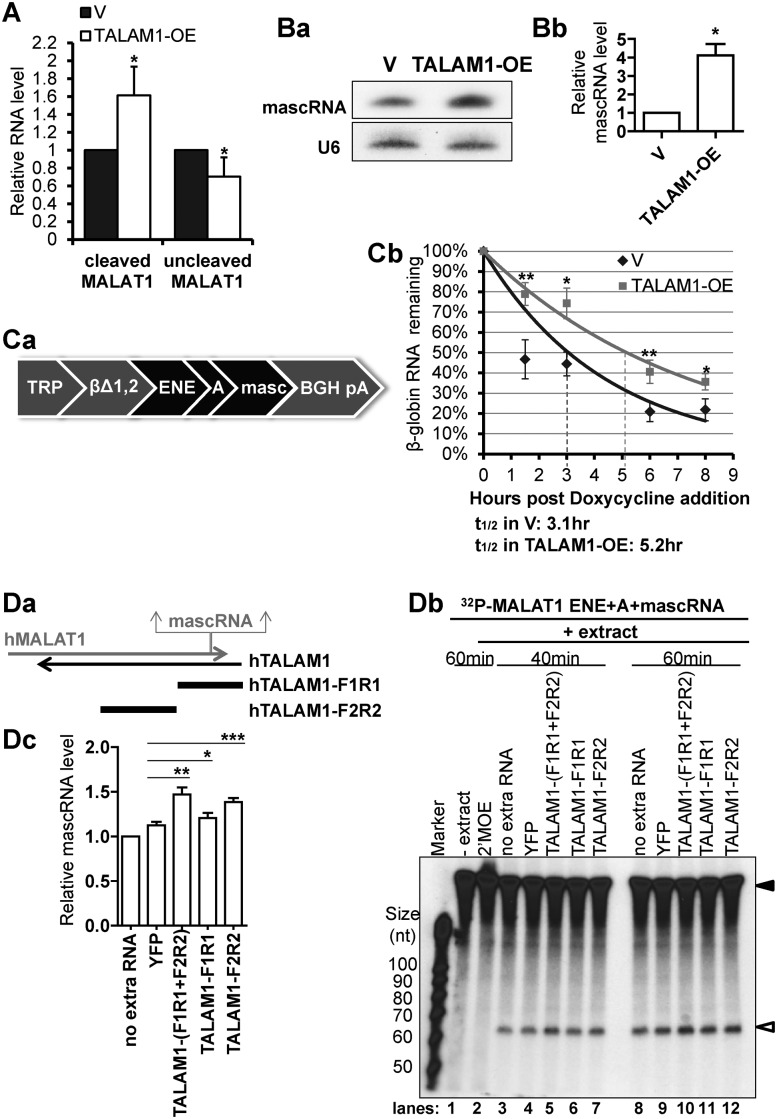
Figure 4.
TALAM1 promotes the 3′ end processing of MALAT1 RNA. (A) Cutoff assay of cleaved and uncleaved MALAT1 in HeLa cells transfected with empty vector (V) or with TALAM1-overexpression construct (TALAM1-OE). (B) Analyses of mascRNA levels in V or TALAM1-OE samples by (Ba) northern blot, and (Bb) Taqman small RNA assay. (C) In vivo RNA decay assay. (Ca) Schematic representation of the βΔ1,2 construct. (Cb) Stability of the intronless β-globin transcript in U2OS Tet-off cells transfected with empty vector (V) or with TALAM1-overexpression construct (TALAM1-OE). Trendlines are single-exponential regression of the percentage of RNA remaining versus time. Dotted lines represent the half-life of reporter RNA. (D) In vitro processing assay of MALAT1 3′ end sequence. Internally 32P-labeled MALAT1 ENE+A+mascRNA RNA substrate (filled arrow in Db) was incubated with HeLa total cell extract in absence or presence of in vitro transcribed YFP, TALAM1-F1R1, F2R2 RNA or 2′MOE targeting the mascRNA site, and its processing to mascRNA (open arrow in Db) is monitored at 40 min and 60 min time points. (Da) Schematic diagrams shown for the relative positions of the TALAM1-F1R1 and TALAM1-F2R2 fragments generated by in vitro transcription. (Db) A representative autoadiogram is shown. (Dc) The quantification of the mascRNA product levels measured from independent experiments, with ImageJ software. At 40 min time point, levels of mascRNA produced under different conditions were normalized to the condition with no extra RNA, which was set as one. Bar data in (A) and (Bb) are mean values with SD, NA = 4, NBb = 3; Data in (Cb) and (Dc) are mean values with SEM, NCb = 4, NDc = 6 (biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed paired Student t-test).