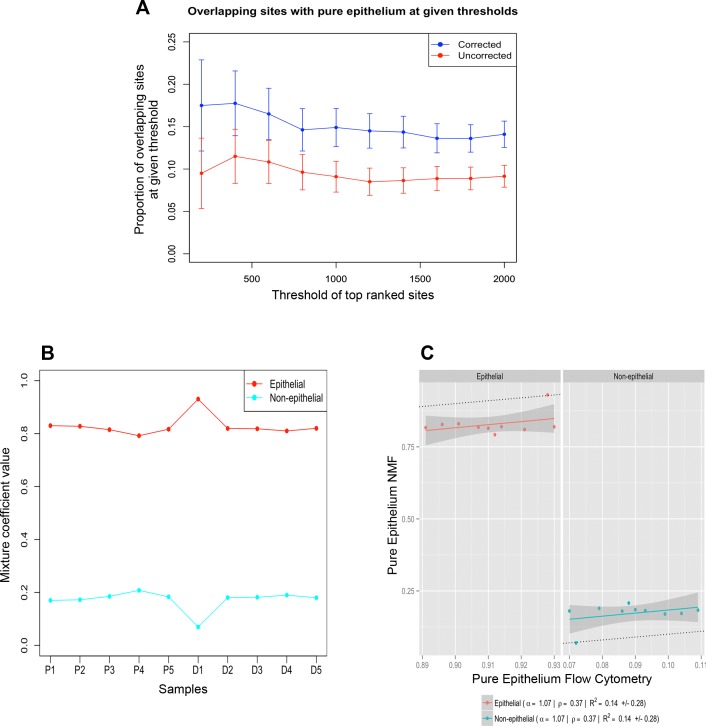
Fig. 7.
Validation of deconvolution accuracy. Overlapping differentially methylated sites (DMS). The proportions of overlapping DMS between whole biopsies (red) and deconvolution corrected whole biopsies (blue) with pure epithelial samples. Lists of DMS identified in each of the 3 cases were ranked according to the mean difference in DNA methylation. For each rank threshold, t, shown on the x-axis, the number of sites in common between the top t sites in the corrected or uncorrected lists and the top t sites identified in the purified epithelium is shown (A). The SEs were constructed based on the upper and lower limits of the estimated confidence interval of the overlap proportions. Profile plot illustrating cell proportion estimates of epithelial (red) and nonepithelial (cyan) populations in whole purified cell samples (B). Scatter plot illustrating the correlation (Pearson = ρ) between measured cell signature estimates of the purified cell samples using flow cytometry (x-axis) and cell signature estimates of the purified samples generated by deconvolution (y-axis) (C).