Figure 2.
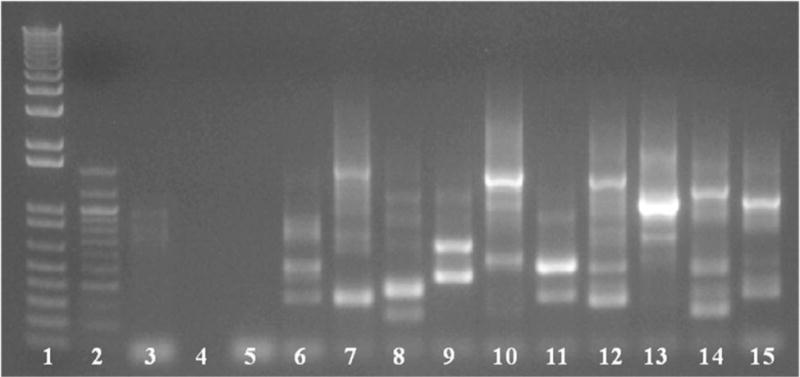
Nested semi-random PCR for sequencing/identification of individual mutants. PCR from individual colonies was carried out using primers specific to the transposon and random primers for the P. gingivalis genome. Agarose gel following second round of nested PCR shown above. Lanes 1 and 2 contain DNA ladder markers, lanes 3 through 5 are negative controls of WT P. gingivalis strain W83, template only and primer only, and individual mutants in the remaining lanes 6–15. Using SAMSeq3 primer for sequencing one will only receive a single product, even though there are multiple bands, unless the prepared gDNA contains two mutant strains. Of note, insertions within the same gene can give different banding patters, as similar banding patterns can be coincidental as well, so extrapolation of insertion location from PCR gel bands is not advisable.
