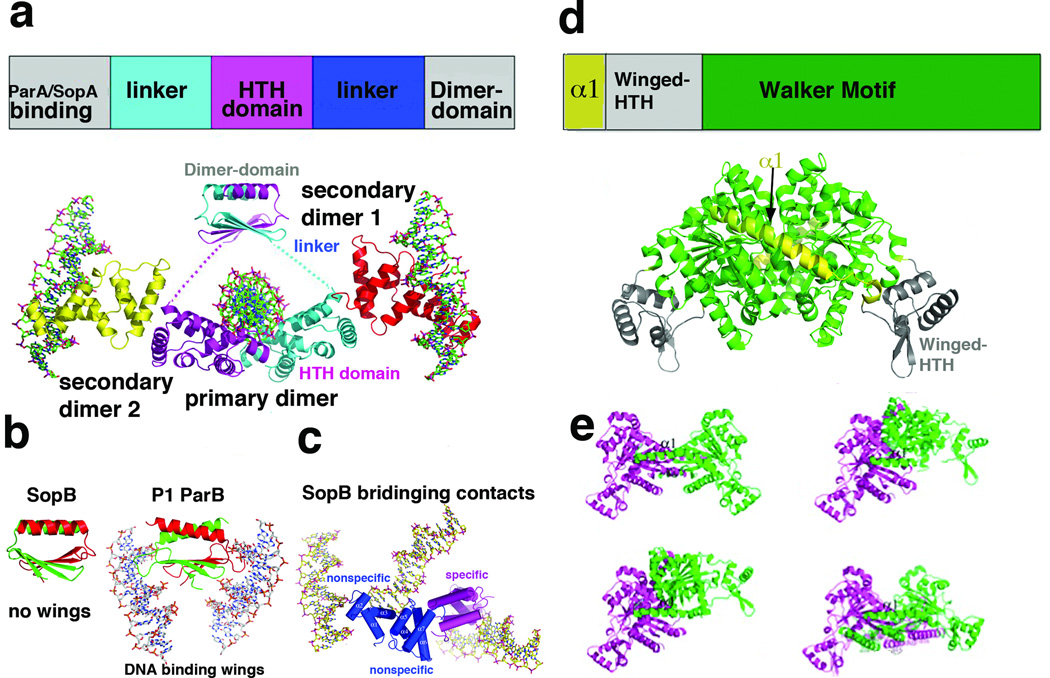
Figure 2.
Structure and domain organization of type Ia partition proteins. (a) Top, schematic diagram showing the domain arrangement of type Ia CBPs. Below is the structure of type Ia CBP SopB showing the dimer-domain and HTH domains, which are flexibly attached [16]. Labeled are the primary dimer (that formed on a single palindromic DNA site) and secondary dimers, which mediate bridging between DNA sites. The four SopB subunits are colored differently. (b) Comparison of the SopB dimer-domain with that of P1 ParB. Although the overall folds of the dimer-domains and their dimerization modes are similar, SopB lacks the wings that are present in P1 ParB, which allow the latter protein to bind DNA [15–16]. (c) SopB-DNA structure in which two nonspecific DNA interactions and one specific interaction was captured. These bridging interactions explain how SopB can spread in trans to surrounding DNA once bound to its centromere. (d) Top, schematic diagram of the type Ia NTPase domain organization as revealed by the P1 and P7 ParA structures [29]. Below the diagram is the P1 ParA structure with domains colored as in the schematic. (e) Structures of four different apo ParA structures with the magenta subunit shown in the same orientation to highlight the dramatic conformational flexibility exhibited by the ParA dimer.