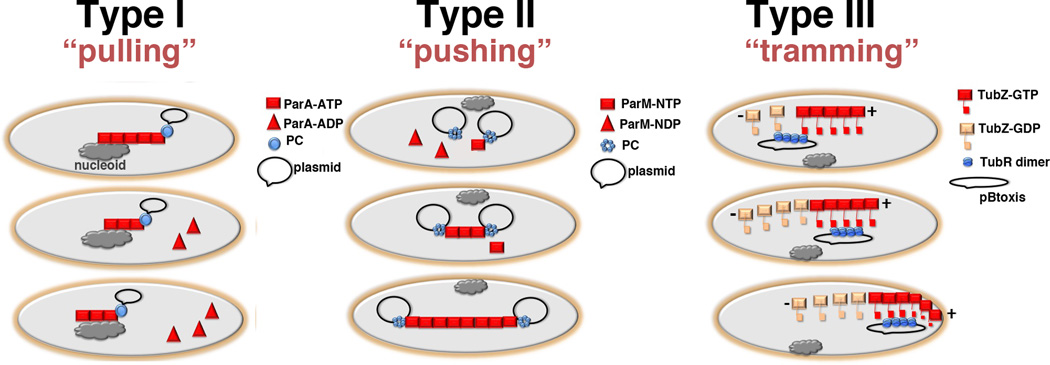
Figure 5.
Schematic models for type I, II and III plasmid partition (a) Type I partition utilizes the host cell nucleoid as a “track” for NTPase-ATP binding and polymerization (square). When the NTPase-ATP polymer encounters a ParB-centromere partition complex (shown as a circle), i.e. the ParB attached plasmid, the NTPase activity is activated resulting in dissociation of capping ParA-ADP subunits (triangles) and polymer retraction. The ParB-plasmid is either pulled along in the retreating ParA polymer or is attracted and diffuses toward the moving polymer. The ultimate outcome is the dynamic equi-distribution of ParB-plasmids at opposite ends of the nucleoid. (b) Type II partition uses a pushing or insertional polymerization mode of segregation. In this model, the dynamically unstable ParM filaments are stabilized and propagate only when each end is captured by a ParR-centromere partition complex. The polymer continues to grow upon addition of ParM-ATP or ParM-GTP subunits to the ParR-ParM interface. The outcome is redistribution of replicated plasmids to opposite poles. (c) Type III partition employs a tram mechanism of partition. TubR binds the centromere serving as a high local concentration of binding sites for the C-terminal flexible domains emanating from treadmilling TubZ filaments. Once captured, the TubR-plasmid is transported to the cell pole by the treadmilling TubZ filaments. Upon reaching the membrane the TubZ filament bends, likely dumping its TubR-plasmid cargo, and reverses direction. Now traveling in the opposite direction, the TubZ filament binds another TubR-plasmid cargo and carries it to the opposite pole.