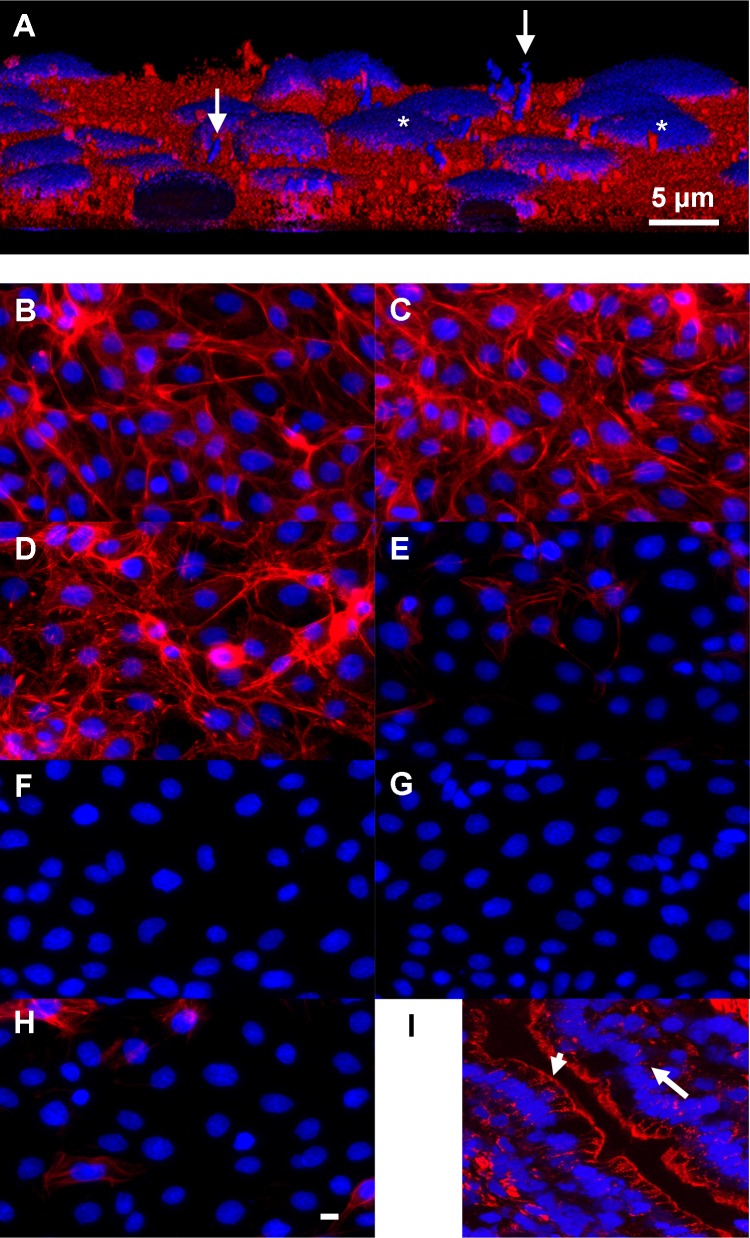
Fig 2. Attachment and structural effects of lactobacilli on IPEC-1 cells.
(A) Three dimensional reconstruction of lactobacilli attached to IPEC-1 cells. Lactobacillus amlylovorus strain GRL1110 (arrows) was applied for 6 h on confluent IPEC-1 cells. The bacterial suspension was then carefully removed; cells and attached bacteria were fixed and stained with DAPI (nuclei, blue) and CM-Dil (plasma membrane, red). Confocal sections were reconstructed to obtain a 3D view. Note the thin, barely visible cytosolic layer above nucleus (*). (B-H) Actin structure of IPEC-1 cells treated with lactate and Lactobacillus amylovorus supernatants. IPEC-1 cells were cultured on glass, incubated as indicated and stained for F-actin (Phalloidin-PromoFluor 546) and nuclei (DAPI). (B) control buffer pH7, (C) Na-DL-lactate (25 mM, pH 7), (D) control buffer pH 4, (E) lactic acid pH 4. (F-H) supernatants of Lactobacillus amylovorus strains. (F) GRL1110, (G) GRL1112, (H) GRL1115. Predominantly basal F-actin fibre structure was reduced by lactic acid and bacterial supernatants (pH 4), (I) F-actin staining of freeze-section of porcine jejunal epithel. Note positive F-actin signal indicating the apical terminal web (arrowhead), the low F-actin signal at the lateral side and the faint signal in the basal region of the enterocytes (arrow). Bar: 5 μm.