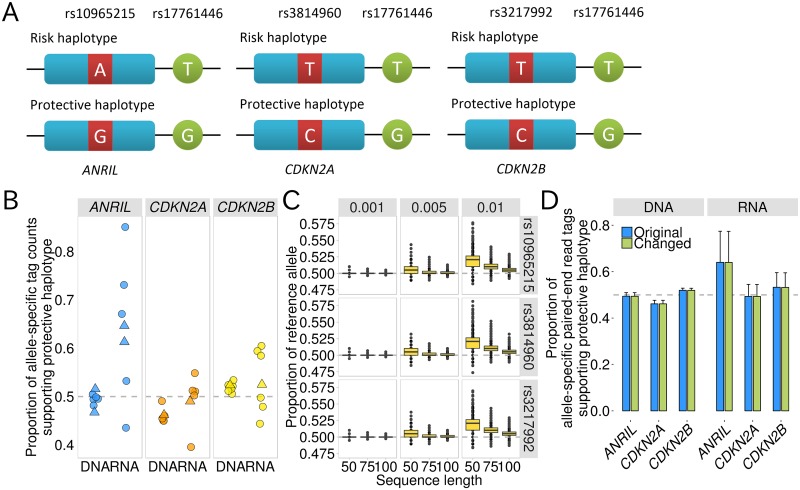
Fig 4. ASE analyses of rs17761446 for 9p21 genes.
A) Haplotypes constructed with candidate causal SNP rs17761446 and SNPs within transcribed region of 9p21 genes (ANRIL, rs10965215; CDKN2A, rs3814960; and CDKN2B, rs3217992). B) Proportions of allele-specific paired-end tag counts supporting the protective haplotype in genome DNA and RNA samples from eutopic endometrial tissues (circles) and endometrial carcinoma cell lines (triangles). C) Simulation study for evaluating mapping bias. Simulation scenarios considering different sequence lengths (50bp, 75bp, and 100bp) and different error rates for each sequenced base (0.01, 0.005, and 0.001). Box plots are based on 10,000 replicates. D) Sensitivity analysis for evaluating mapping bias. In sensitivity analyses, we aligned reads from ASE to two types of sequences that were different in the base at the SNP site: the “original” and “changed” sequences have “reference” and “alternative” alleles, respectively. For each bar plot, mean and standard deviation are shown. Note that the significant result from ANRIL exhibited increased proportion of alternative allele.