Figure 2.
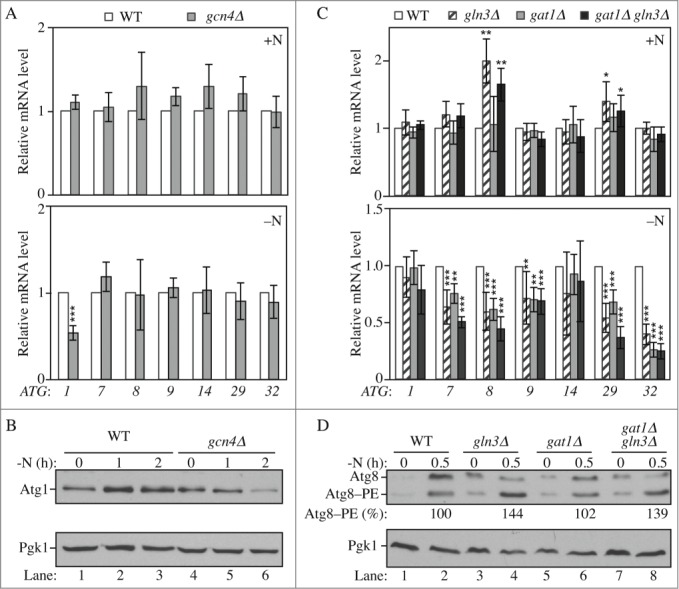
Gln3, Gat1 and Gcn4 are transcriptional modulators of ATG genes upon nitrogen starvation. (A-B) Gcn4 is required for the proper induction of ATG1 upon nitrogen starvation. (A) Wild-type (WLY176; SEY6210) and gcn4Δ (YAB387) cells were grown in YPD (+N) until mid-log phase (upper panel) and then starved for nitrogen (-N) for 1 h (lower panel). mRNA levels were quantified by RT-qPCR. The mRNA level of individual ATG genes was normalized to the mRNA level of the corresponding gene in wild-type cells, which was set to 1. The data represent the average of at least 3 independent experiments. (B) Wild-type and gcn4Δ cells were grown in YPD until mid-log phase and then starved for nitrogen (-N) for the indicated times. Protein extracts were analyzed by western blot with anti-Atg1 and anti-Pgk1 (loading control) antisera. (C-D) Gln3 and Gat1 are required for the proper induction of ATG7, ATG8, ATG9, ATG29 and ATG32 after nitrogen starvation; the deletion of GLN3 increases the expression of ATG8 and ATG29 in growing conditions. (C) Wild-type (WLY176), gln3Δ (YAB385), gat1Δ (YAB384) and gat1Δ gln3Δ (YAB386) cells were grown and mRNA analyzed as in (A). The data represent the average of at least 3 independent experiments. (D) For the analysis of Atg8 protein level, wild-type (WLY176; SEY6210), gln3Δ (YAB385), gat1Δ (YAB384) and gat1Δ gln3Δ (YAB386) cells were grown as in (B). Protein extracts were analyzed by western blot with anti-Atg8 and anti-Pgk1 (loading control) antisera. The percent Atg8–PE of total Atg8 is indicated.
