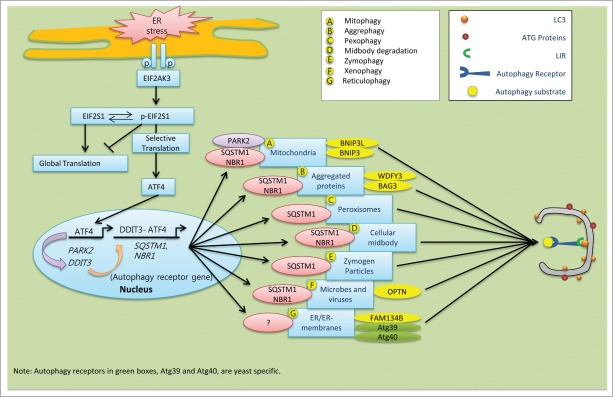
Figure 2.
Schematic model of the speculative roles of ER stress components involved in selective autophagy in mammalian cells. During ER stress, EIF2AK3-dependent ATF4 translation and its interaction with DDIT3 in the nucleus stimulate expression of the autophagy receptor genes SQSTM1 and NBR1, which are ubiquitous. (A) ER-stress-mediated activation of ATF4 sustains the expression of PARK2. Damaged mitochondria recruit PARK2 as a degradation signal recognized by various autophagy receptors, including SQSTM1 and NBR1. (B) Following ER stress, upregulated SQSTM1 and NBR1 interacts with ubiquitin aggregates and enhance its selective clearance through aggrephagy. Autophagy receptor WDFY3 and BAG3 are also involved in aggrephagy.185 (C) During pexophagy, SQSTM1 causes superfluous or damaged peroxisomes to cluster and subsequent be delivered into the growing selective phagophore.220 (D) After cell division, midbody remnants are recognized by the autophagy receptors SQSTM1 and NBR1, which deliver them to the phagophore for degradation.221 (E) The ubiquitin-binding protein SQSTM1 interacts with and traffics ubiquitinated zymogen granules to the nucleating phagophores.222 (F) Ubiquitinated proteins interact with OPTN. The resulting autophagy receptor complex accelerates the selective autophagy flux of viruses, microbes, and other non-host entities.223 (G) In mammals, the autophagy receptor FAM134B interacts with LC3 and GABARAP, and facilitates ER turnover by reticulophagy.175 Similarly, yeast specific reticulophagy is Atg39 and Atg40 dependent.224,225 However, the role of SQSTM1 and NBR1 in reticulophagy has not been clarified yet.