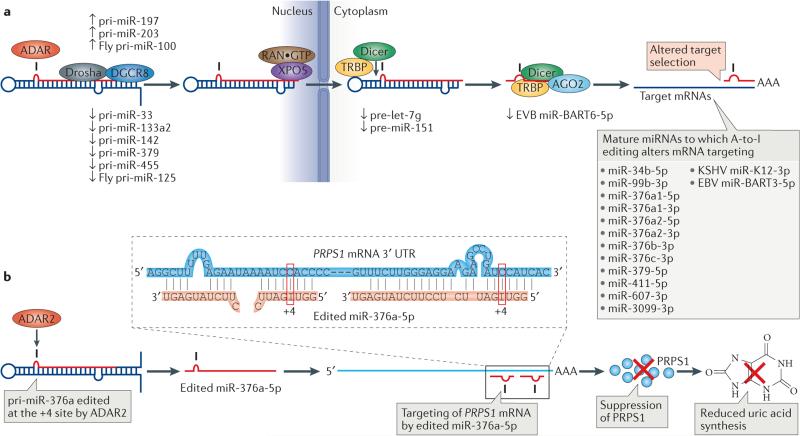
Figure 4. Regulation of microRNA (miRNA) processing, expression and selectivity by RNA editing.
a | Primary miRNAs (pre-miRNAs) are processed by the Drosha–DGCR8 complex into precursor miRNAs (pri-miRNAs) in the nucleus, exported to the cytoplasm by exportin 5 (XPO5)–RAN·GTP and processed by the Dicer–TAR RNA-binding protein (TRBP) complex into mature miRNA duplexes. One strand of this duplex is then loaded onto the RNA-induced silencing complex (RISC), which results in the degradation or the inhibition of translation of target mRNAs. Editing can affect any of the miRNA biogenesis steps, including Drosha cleavage, Dicer cleavage and RISC loading, as well as miRNA target selection. Known examples of miRNA editing and their consequences are shown. b | Silencing of phosphoribosyl pyrophosphate synthetase 1 (PRPS1) by miR-376a-5p edited at the +4 site by ADAR2 (adenosine deaminases acting on RNA2) and the consequent suppression of uric acid synthesis. A single A-to-I nucleotide change in the seed sequence of miR-376a-5p results in redirection of target gene selection. One of those genes, specifically targeted by the edited miR-376a-5p, is PRPS1, which encodes an essential enzyme involved in purine metabolism and the uric acid synthesis pathway. Repression of PRPS1 by the edited miR-376a-5p results in reduced expression of uric acid in certain tissues, such as brain, in which uric acid levels need to be tightly regulated. The 3′ untranslated region (UTR) of PRPS1 mRNA has two target sites for the edited miR-376a-5p (inset). AGO2, Argonaute 2; EBV, Epstein–Barr virus; KSHV, Kaposi sarcoma-associated herpes virus.