FIGURE 1.
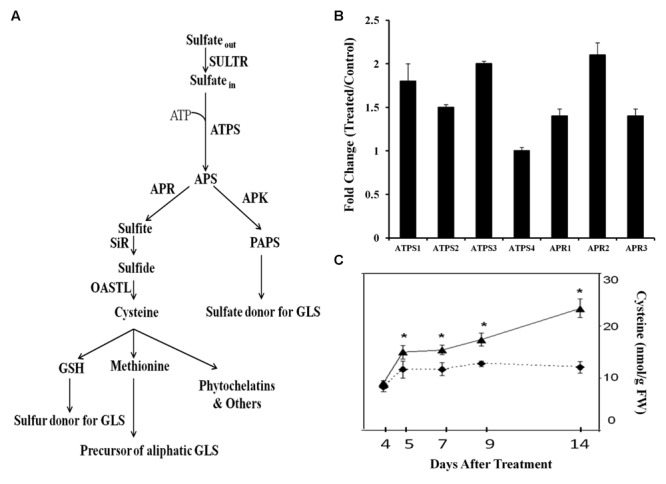
Transcriptional regulation of Arabidopsis sulfur assimilation genes by GB03. Depicted sulfate assimilation pathway adapted from Mugford et al. (2009) (A). Semi-quantitative RT-PCR analysis of whole-plant sulfur assimilation gene expression at 72 h post GB03 treatment (B); data are the averages of three biological replicates with error bars representing standard error. The amino acid cysteine increases with GB03 treatment (solid line) relative to the water controls (dashed lines; C); an asterisk (∗) indicate statistically significant difference between treatments (t-test, P-value ≤ 0.05, n = 6, mean ± SE). Sulfur assimilation pathway includes SULTR, sulfate transporter; ATPS, ATP sulfurlyase; APS, adenosine 5′-phosphosulfate; APR, APS reductase; SiR, sulfite reductase; OASTL; O-acetylserine (thiol) lyase; GSH, glutathione; APK, APS kinase; PAPS, 3′-phosphoadenosine 5′-phosphosulfate.
