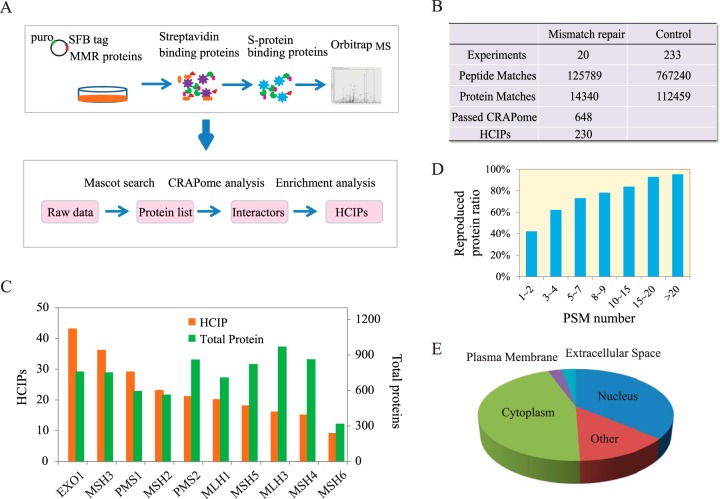
Fig. 1.
Proteomics analysis of human DNA mismatch repair pathway. (A) Diagrams of interactome study pipeline using tandem affinity purification-mass spectrometry (TAP-MS) approach and HCIP filtration employed in this study. (B) Summary of TAP-MS results of MMR proteins and control experiments, including identified peptides, proteins, and HCIPs. The CRAPome score FC-B > 2 was used as the cutoff to identify HCIPs. Additional filtration using proteomic profile of HEK293T cell was also applied. (C) Total numbers of identified proteins and corresponding HCIPs for each DNA MMR protein were presented. (D) The data reproducibility distribution within TAP-MS experiment repeats. As expected, proteins with higher numbers of PSMs have higher reproducibility. (E) The localization analysis of HCIPs of DNA MMR proteins is presented.