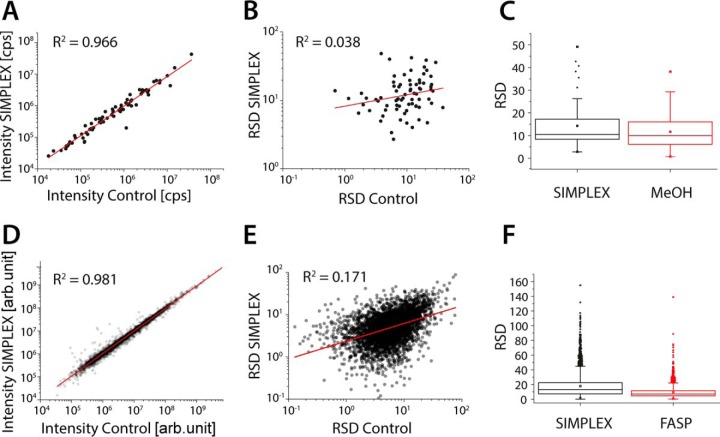
Fig. 2.
SIMPLEX is as efficient as state-of-the-art protocols for metabolomics and proteomics. To compare SIMPLEX against other extraction protocols the proteins were extracted with and without MeOH precipitation prior FASP, followed by a standard proteomics workflow. For metabolites, a standard MeOH extraction versus the MTBE/MeOH extraction was used to demonstrate the comparability of the proposed strategy. Correlation plots and linear fits of the identified molecular features (A) metabolites (control versus SIMPLEX) and (D) proteins (control versus FASP). To investigate if the error between the samples is correlating, the relative standard deviation (RSD) was plotted for each identified feature against the S.D. of the corresponding feature from the control experiment for metabolites (B) and proteins (E). (C) and (F), displaying the average, median RSD for metabolites and proteins for the SIMPLEX and control experiment as well as their error distribution (n = 3 for all experiments).