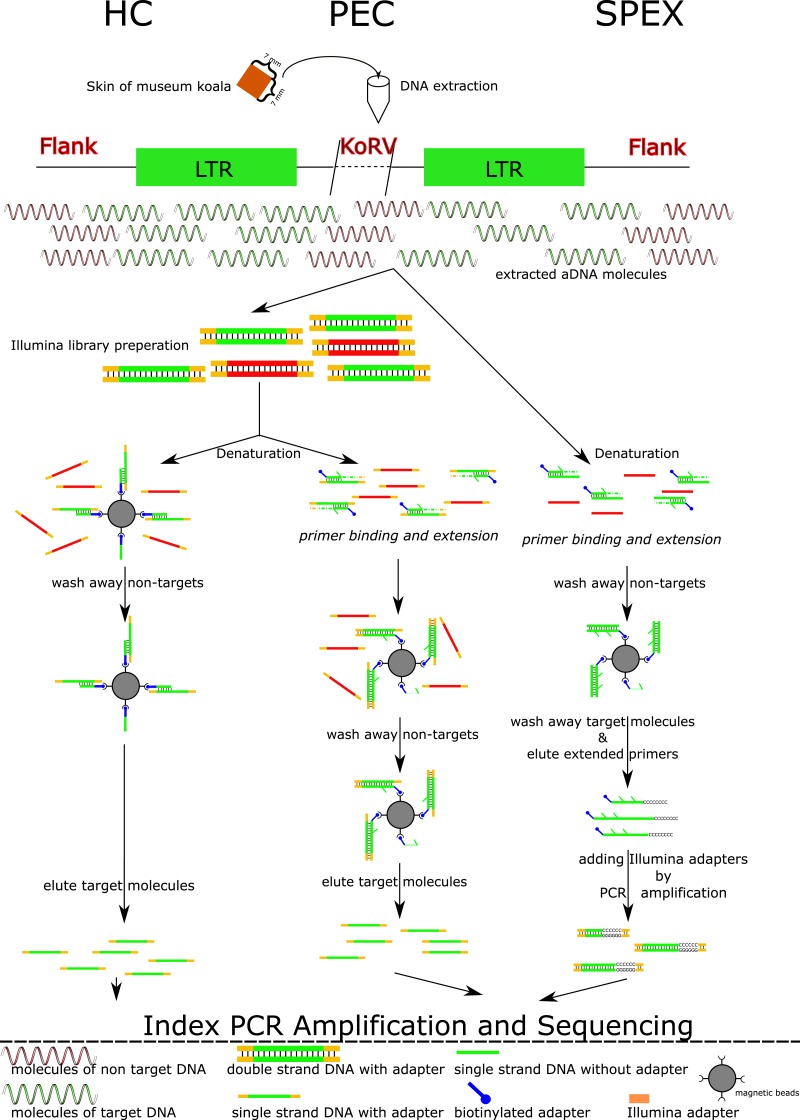
Figure 1. Experimental work flow for the three enrichment techniques.
Abbreviations: HC, Hybridization Capture; PEC, Primer Extension Capture; SPEX, Single Primer Extension. A square of 7 mm × 7 mm of koala skin tissue per museum specimen was extracted in a dedicated ancient DNA (aDNA) facility. A workflow for the three techniques is illustrated. Both HC and PEC require Illumina library preparation as a preliminary step. The double stranded libraries are denatured to single stranded DNA molecules and underwent different experimental procedures in HC and PEC. In HC, single stranded DNA libraries are mixed with magnetic beads immobilized with baits. These are incubated by slow rotation at 65 °C for 48 h. After a series of wash steps, the libraries with non-targets sequences are washed off leaving only the libraries with target sequences hybridized with the baits on beads. These target molecules were then dissociated from the baits using a special elution buffer, and were used as templates for PCR amplification. While in PEC, the singled stranded libraries are mixed with biotinylated oligos for 1 min at 55 °C in which only the libraries with target sequences hybridized with the biotinylated oligos. Primer extension reactions of the biotinylated oligos were performed only to these hybridized libraries. Biotinylated oligos were collected by magnetic beads together with the hybridized targeted libraries. The single stranded libraries with target sequences were dissociated with biotinylated oligos and were eluted for subsequent PCR amplification. In contrast for SPEX, DNA extracts are directly denatured to be single stranded and mixed with the same biotinylated oligos used in PEC for 1 min at 55 °C. Similar as in PEC, primer extension reactions of the biotinylated oligos were performed only to the single molecules (target sequences) hybridized with biotinylated oligos. These hybrid molecules were collected using magnetic beads. The original single stranded target molecules were washed away and the biotinylated oligos with 3′ extension were eluted off the beads and were treated with a poly C tailing reaction. These poly C tailed molecules were amplified using primers with a 5′ overhang of the Illumina sequencing adaptor. Through this process, the SPEX products were constructed into Illumina libraries without an additional library preparation step. These SPEX-Illumina libraries were then used in an index PCR and a further amplification step. As shown, SPEX requires at least one more amplification step than HC or PEC, which may explain the high level of clonality in the SPEX result.