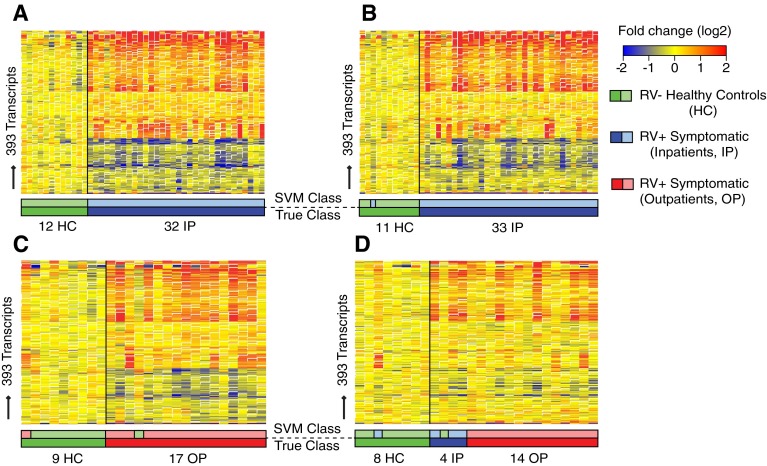
Figure 2.
Symptomatic rhinovirus (RV) infection induces a robust and reproducible transcriptional signature. (A) Training set (n = 44). Class comparisons (Benjamini-Hochberg corrected false discovery rate <0.01 and ≥1.25-fold change) using linear models adjusted for age, race, and white blood cell composition (neutrophil score) between RV− healthy control subjects (HC) and RV+ inpatients (IP) identified 393 differentially expressed transcripts (transcriptional RV signature) (Table E4). Transcripts are organized in a heat map format in which each row represents a single transcript and each column represents a patient sample. Red indicates overexpression, and blue underexpression, of a transcript compared with the median expression of HC (yellow). The transcriptional signature was validated in three independent patient sets using the support vector machine (SVM) algorithm. The SVM algorithm predicted the condition (symptomatic RV infection or RV− HC) in (B) the test set (n = 44) with 98% accuracy and in (C) validation set A (n = 26) and (D) validation set B (n = 26) with 92% accuracy. Color bars below the heat maps indicate the true sample class below (darker colors) and above (lighter colors) the predicted SVM class. OP = outpatient.