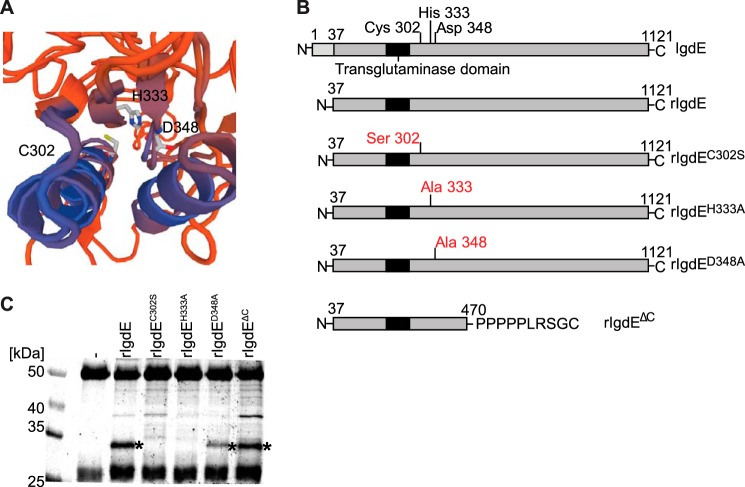
FIGURE 2.
Identification of the active site of IgdE. A, in silico modeling of IgdE reveals Cys-302, His-333, and Asp-348 as putative catalytic triad residues. B, schematic illustration of IgdE and the different rIgdE constructs with potential active site residue substitutions and the C-terminal truncation variant. The secretion signal peptide (residues 1–37) is marked in light gray, the transglutaminase domain is boxed, and the potential active site residues and substitutions are indicated. C, 3.3 μm porcine IgG was incubated for 16 h at 37 °C with soluble fractions of E. coli cells expressing different rIgdE constructs. The reactions were analyzed by SDS-PAGE under reducing conditions. IgG cleavage (*) occurred upon incubation with rIgdE, rIgdED348A, and rIgdEΔC but not with rIgdEC302S or rIgdEH333A. The weak protein band of 37 kDa is a contaminant present in lysate preparation and not related to IgdE activity.