FIGURE 4.
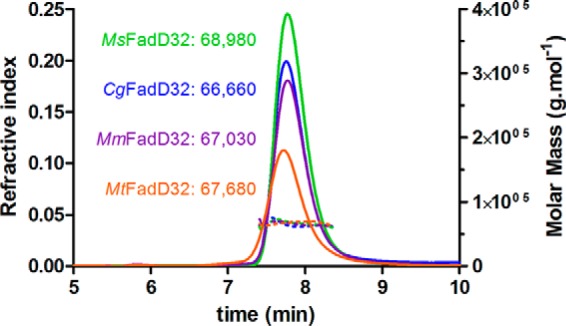
SEC-MALS experiments on FadD32 enzymes. Continuous lines represent the variation of refractive index against elution time from the size exclusion column for MtFadD32 (orange), MmFadD32 (purple), MsFadD32 (green), and CgFadD32 (blue). The experimentally measured molar mass distribution (dashed lines, same color code) and deduced mean molar mass obtained with ASTRA software (in g·mol−1) are indicated for each elution peak. Calculated molecular masses of untagged proteins used for experiments (in daltons): MtFadD32, 69,513; MmFadD32, 69,172; MsFadD32, 68,510; CgFadD32, 68,196. At the flow rate used (0.35 ml·min−1), the exclusion limit of the column corresponds to an elution time of 5.8 min.
