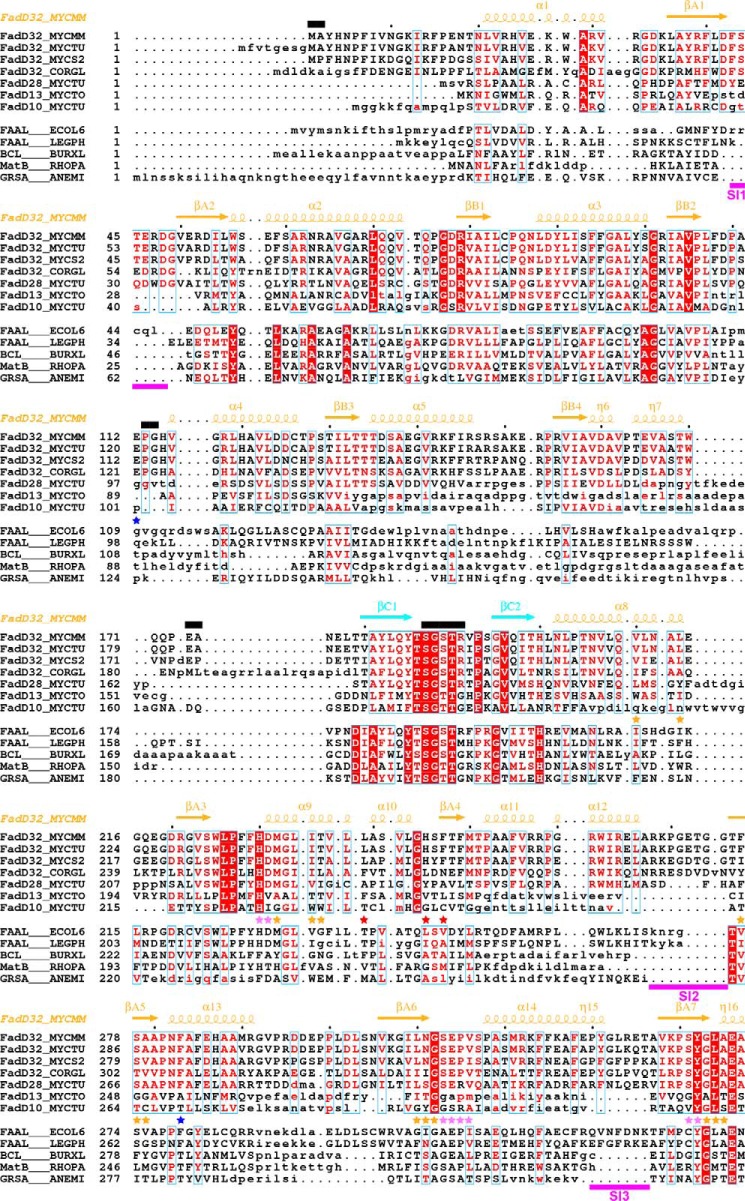
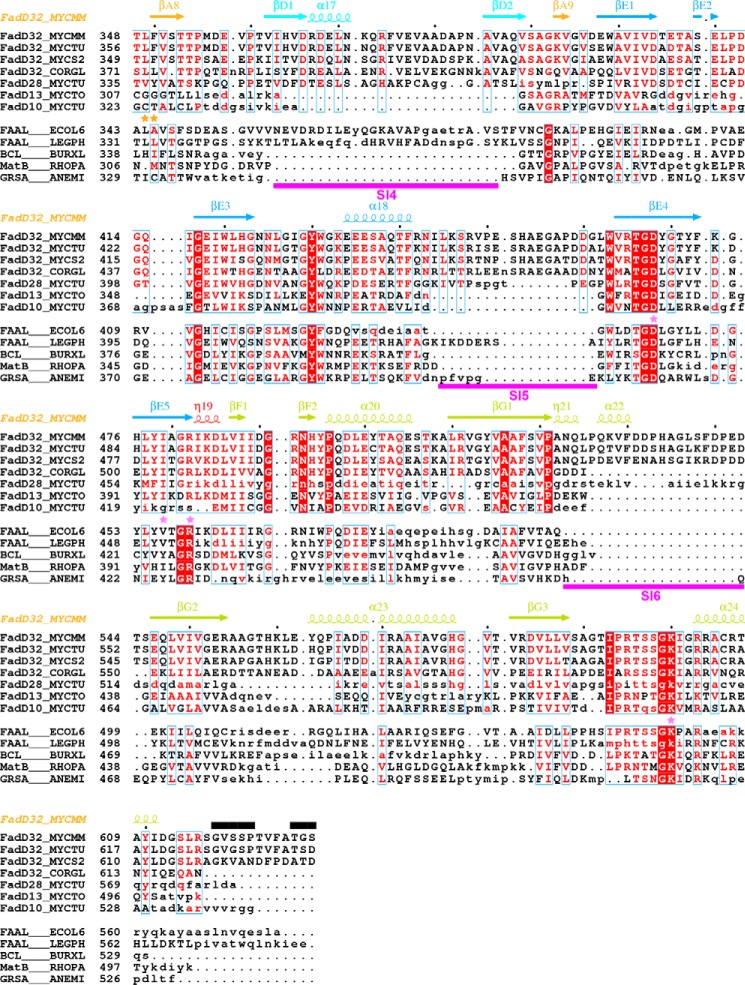
FIGURE 6.
Structure-based alignment of the sequences of mycobacterial FadDs with those of other adenylate-forming enzymes. Sequences were separated in two groups (upper group, selected mycobacterial FadDs; lower group, selected adenylate-forming enzymes). Within each group, sequence similarity is indicated by red letters, whereas sequence identity is indicated by white letters on a red background. Aligned and unaligned residues are displayed in uppercase and lowercase, respectively, taking MmFadD32 as reference. Residues 460–580 of FadD28, also in lowercase, are absent from the structure and were aligned manually. Secondary structure elements (arrows for β-strands and coils for α- and η-helices) of MmFadD32 are indicated at the top. Residues of MmFadD32 that are disordered in the crystal structure are also indicated at the top, by black bars. Sequence insertions (SI1 to SI6) in FadD32 are underlined in magenta. Residues important for alkyl adenylate binding are indicated by violet (adenine moiety) and orange (aliphatic chain) stars. Residues with side chains that undergo conformational changes to accommodate AMPC20 binding are indicated by red stars. Mutations that have been shown to confer resistance to coumarin inhibitors are indicated by blue stars.